Latest posts
-
Play Foldit to help stop coronavirus (VIDEO)
You don’t have to be a scientist to do science! By playing the computer game Foldit, you can help discover new antiviral drugs that might stop the novel coronavirus. The most promising solutions will be manufactured and tested at the University of Washington Institute for Protein Design in Seattle. Foldit is run by academic…
-
Takeda acquires PvP Biologics
PvP Biologics is on a mission to develop a highly-effective therapeutic to reduce the burden of living with celiac disease. They are advancing an oral enzyme — TAK-062 — designed to break down gluten in the stomach. This exciting research, which began as an iGEM project in 2011, was matured…
-
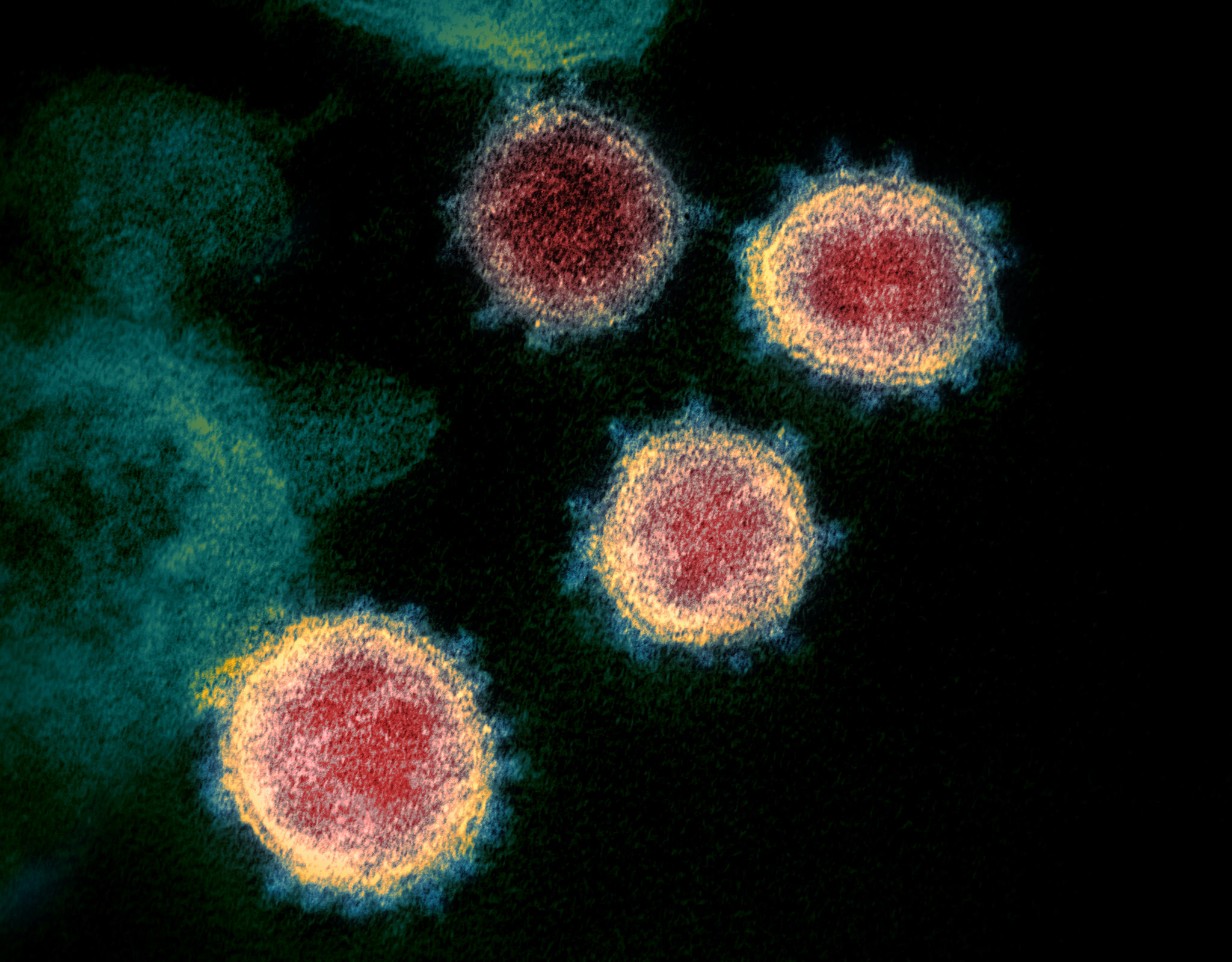
Rosetta’s role in fighting coronavirus
We are happy to report that the Rosetta molecular modeling suite was recently used to accurately predict the atomic-scale structure of an important coronavirus protein weeks before it could be measured in the lab. Knowledge gained from studying this viral protein is now being used to guide the design of…
-
Introducing WE-REACH, a new biomedical innovation hub
With $4 million in matching funds from the National Institutes of Health, the University of Washington has created a new integrated center to match biomedical discoveries with the resources needed to bring innovative products to the public and improve health. “The University of Washington and regional partner institutions produce some…
-
SCI-STEM Symposium 2020
The Institute for Protein Design at the University of Washington held the second symposium aimed at providing strategies to address diversity challenges in science, technology, engineering, and math (STEM). The Strategies for Cultivating Inclusion in STEM (SCI-STEM) symposium featured leading keynote speakers, panel discussions, and interactive breakout sessions. Members of…
-
Radhika wins poster award at ABRCMS
Undergraduate researcher Radhika Dalal took home a poster award at this year’s Annual Biomedical Research Conference for Minority Students in Anaheim, California. Her mentors include IPD graduate student Una Natterman and Quinton Dowling. Way to go, Radhika! And congratulations to all the award winners:
-
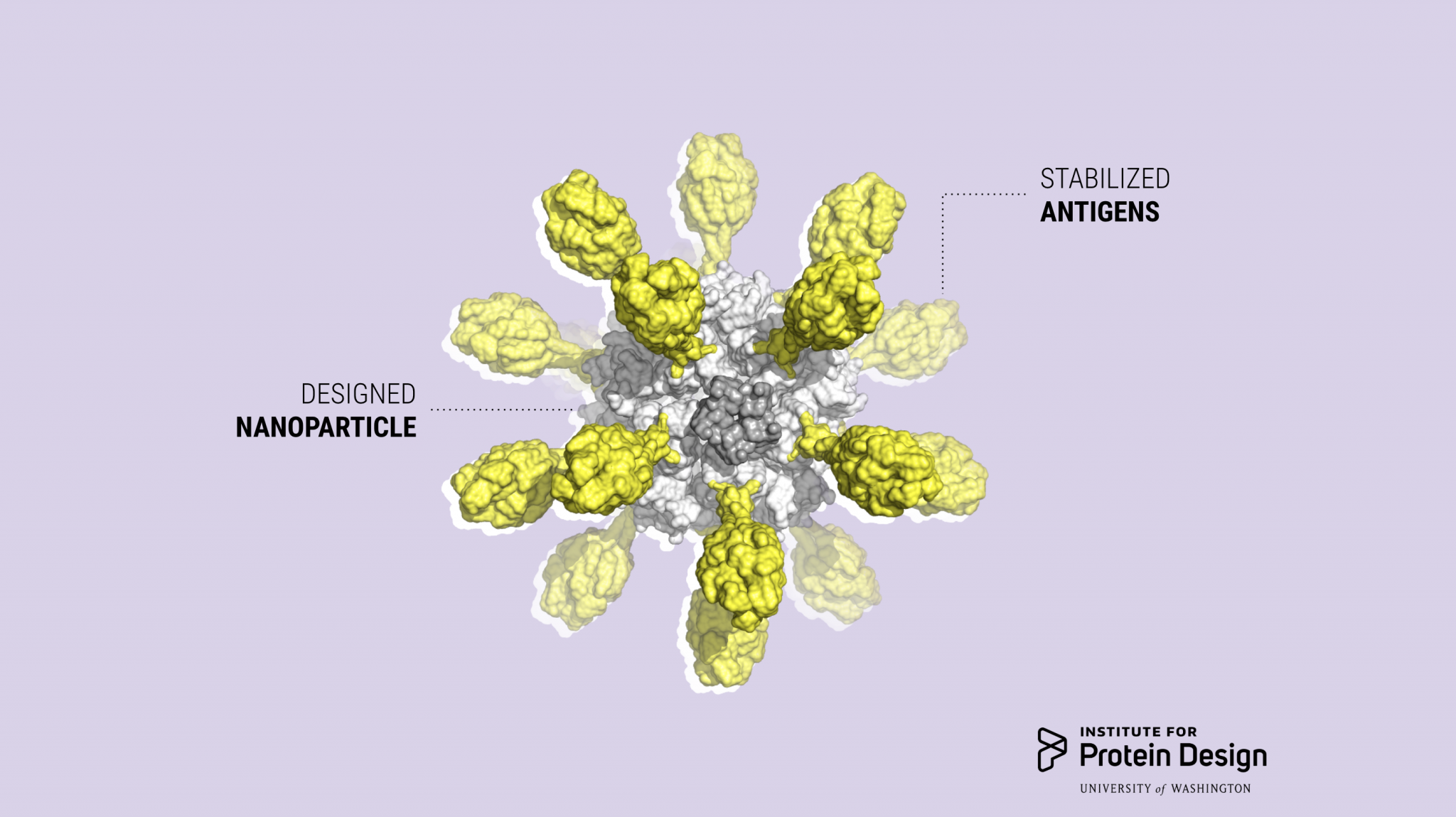
Icosavax launches to advance designer vaccines
Icosavax, Inc. today announced its launch with a $51 million Series A financing. The company was founded on computationally designed self-assembling virus-like particle (VLP) technology developed here at the IPD (Cell 2019, Preview). The proceeds of the financing will be used to advance the company’s first vaccine candidate, IVX-121, for…
-
Neoleukin: from spinout to public company in 7 months
IPD-spinout Neoleukin Therapeutics announced this week a merger with Aquinox Pharmaceuticals, a publicly traded company. The combined company will change its name to Neoleukin Therapeutics, and will continue to advance its Rosetta-designed protein platform for cancer, inflammation, and autoimmune diseases. Neoleukin was spun out of the IPD Translational Investigator Program…
-
5 questions about LOCKR from our Reddit AMA
Researchers from the IPD and UCSF recently participated in a Reddit Ask Me Anything about LOCKR, our new de novo protein switch. Reddit users had dozens of fantastic questions — so many, in fact, that the team ran out of time before they could address them all. “The questions were…
-
Introducing LOCKR: a bioactive protein switch
Today we report in Nature the design and initial applications of the first completely artificial protein switch that can work inside living cells to modify—or even commandeer—the cell’s complex internal circuitry. The switch is dubbed LOCKR, short for Latching, Orthogonal Cage/Key pRotein. “In the same way that integrated circuits enabled the explosion of the computer chip…