Latest posts
-
SCI-STEM Symposium 2018
Update 2018-07-26: The 2018 SCI-STEM Symposium was recently featured in an eLife article. The Institute for Protein Design at the University of Washington held its first ever symposium aimed at providing strategies to address diversity challenges in science, technology, engineering, and math (STEM). The Strategies for Cultivating Inclusion in STEM…
-
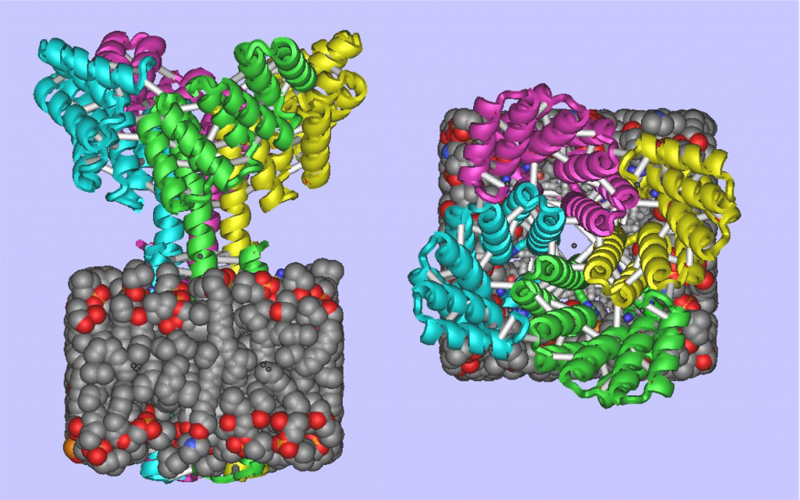
De Novo Design of Membrane Proteins
It is now possible to create complex, custom-designed transmembrane proteins from scratch ! Today Baker lab members published in Science “Accurate computational design of multipass transmembrane proteins” The Abstract reads as follows: The computational design of transmembrane proteins with more than one membrane-spanning region remains a major challenge. We report…
-
David Baker profiled in The New York Times
At the end of a historic year for protein design, the Baker Lab was honored to be profiled in the New York Times by science writer Carl Zimmer. He writes about the technology, progress, and promise in the field, including the contributions from our wonderful crowdsource participants. Graphic: John Hersey /…
-
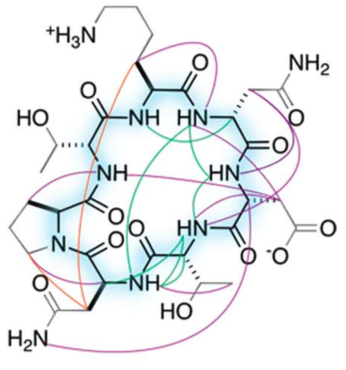
A New World of Designed Macrocycles
Today marks another major step forward for peptide based drug discovery. IPD researchers report in Science the computational design of a new world of small cyclic peptides, “Macrocycles”, increasing the number of the known kinds of these molecules by multiple fold. The conceptual art image below “Illuminating the energy landscape” shows…
-
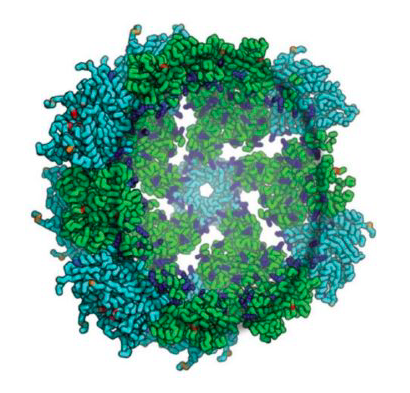
Synthetic Nucleocapsids Have Arrived
Published today in Nature, IPD researchers describe the first synthetic protein assemblies — dubbed synthetic nucleocapsids — that encapsulate their own genome and evolve in complex environments. Synthetic nucleocapsids are built to resemble viral capsids and could be used in future to deliver therapeutics to specific cells and tissues. These icosahedral protein…
-
Foldit to Disarm a Fungal Toxin
Foldit alfatoxin project update 7/16/2018 Today, scientists from of the Institute for Protein Design will join Foldit gamers from around the world to help design an enzyme that can neutralize aflatoxin — a cancer-causing toxin produced by certain fungi that are found on agricultural crops such as corn, peanuts, cottonseed, and tree…
-
Designs on New World of Mini-Protein Therapeutics
Mark your calendars! September 27, 2017 is the day the doors opened to whole new world of targeted therapeutics. The Baker lab and numerous talented collaborators published in Nature that it is now possible to conduct “Massively parallel de novo protein design for targeted therapeutics”. Three factors make this possible: Rosetta molecular…
-
Cyrus Raises $8M to Advance Cloud-Based Protein Modeling and Design
Today, the first IPD spin out company Cyrus Biotechnology announced the closing of an $8M total Series A financing. The investment was led by Trinity Ventures, with participation from OrbiMed Advisors, SpringRock Ventures, W Fund, WRF Capital (a major supporter of the IPD), and individual investors. Congratulations Cyrus team! Cyrus is commercializing Cyrus Bench®…
-
Arzeda Raises $12M for Computational Protein Design
Today, Baker lab spin out company Arzeda announced that it had raised $12 million in a Series A round of funding led by OS Fund and including Bioeconomy Capital and Sustainable Conversion Ventures, as well as a follow-on investment from Arzeda’s seed investor, WRF Capital (a major supporter of the IPD). The…