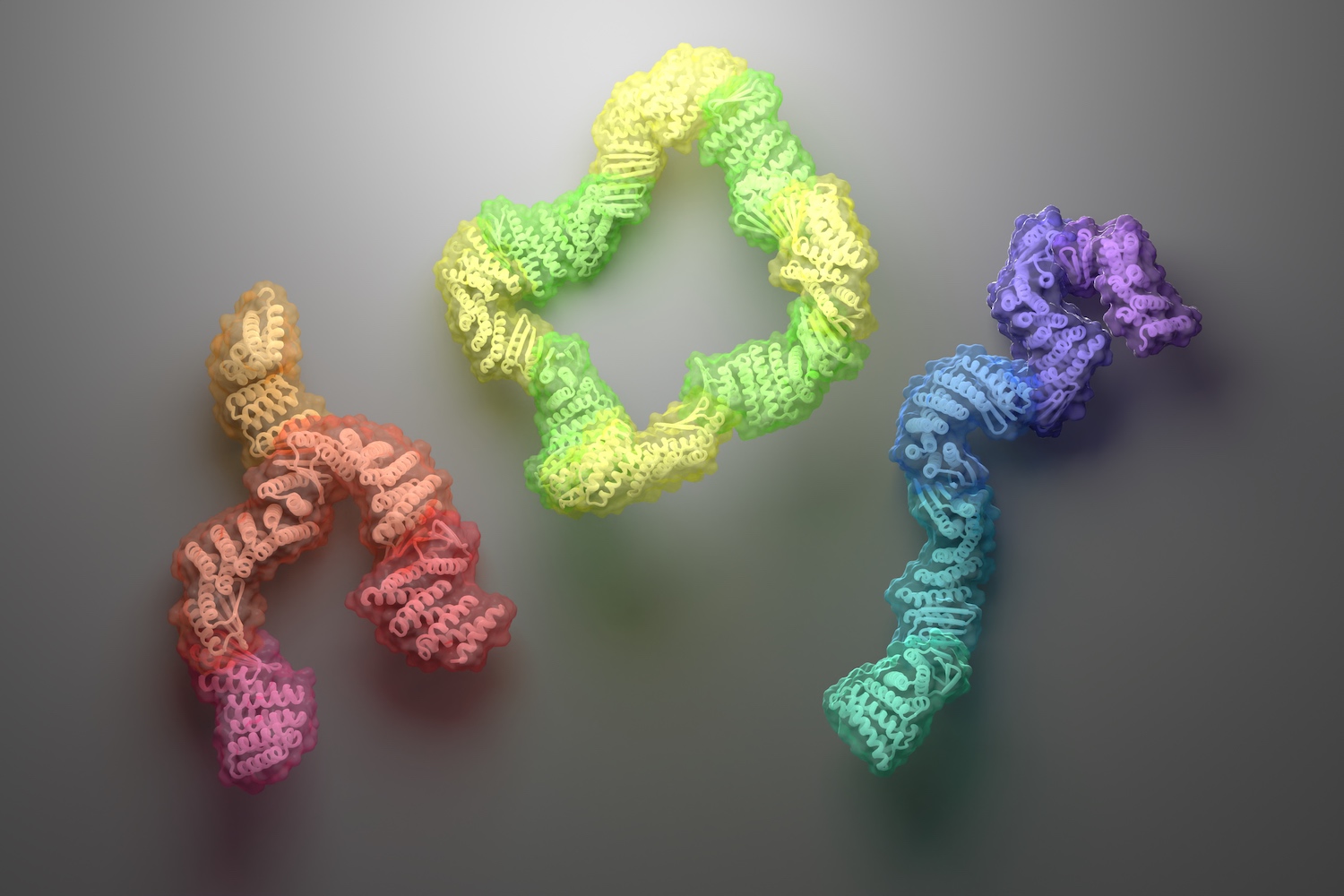Latest posts
-
Anindya Roy wins Go-To-Market Award for drug development
The Washington Entrepreneurial Research Evaluation and Commercialization Hub (WE-REACH) is pleased to announce a product concept award for Dr. Anindya Roy and his team at the UW Medicine Institute for Protein Design, including Drs. Jake Kraft and Hua Bai. They are developing a novel binder protein in an aerosolized delivery…
-
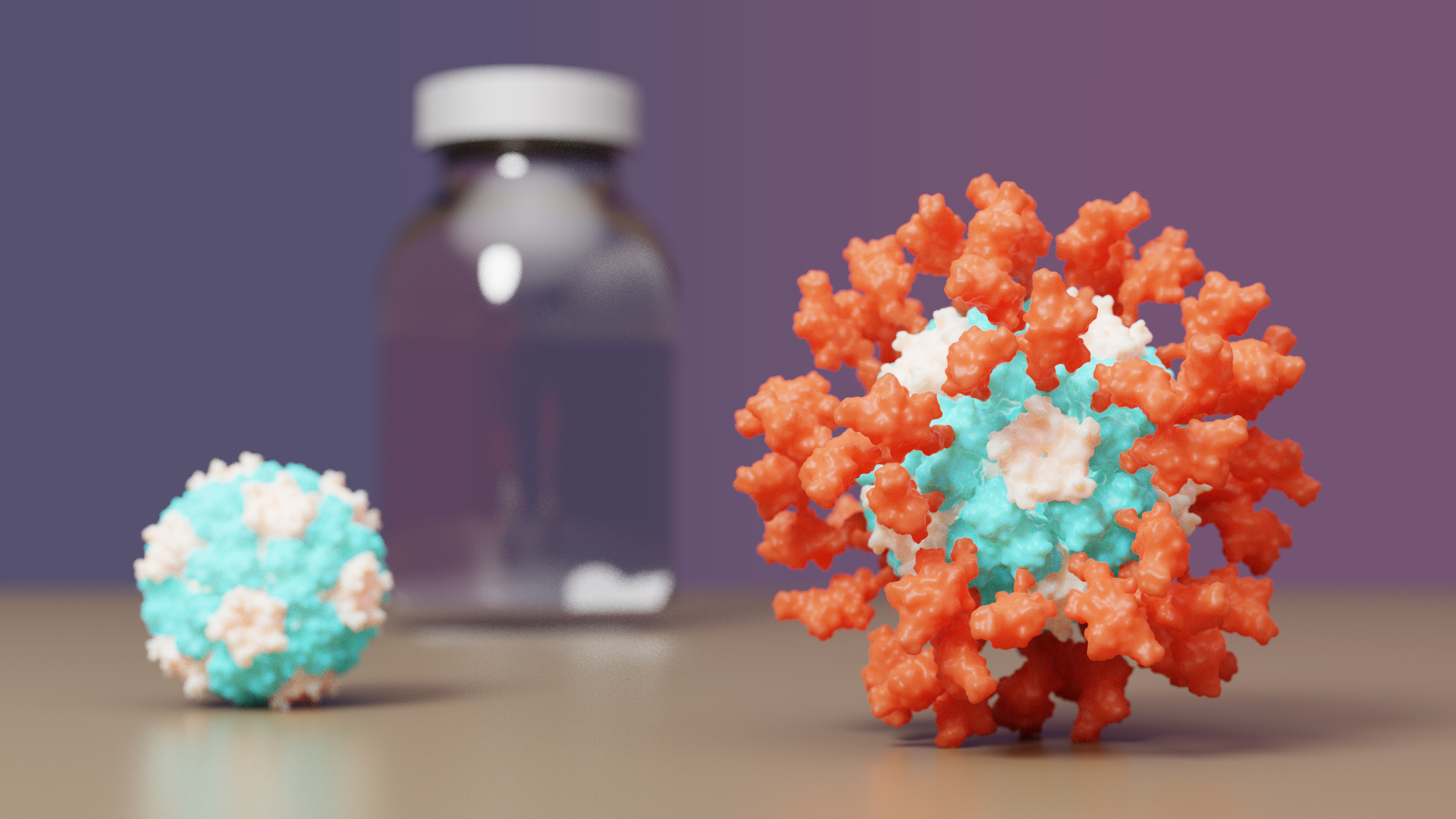
COVID-19 vaccine with IPD nanoparticles seeks full approval
A COVID-19 vaccine developed at the University of Washington School of Medicine has proven safe and effective in late-stage clinical testing. SK bioscience, the company leading the vaccine’s clinical development abroad, is seeking full approval for its use in South Korea and beyond. If approved by regulators, the vaccine will…
-
Custom biosensors for detecting coronavirus antibodies in blood
Today we report in Nature Biotechnology the design of custom protein-based biosensors that can detect coronavirus-neutralizing antibodies in blood. This research, which builds on prior sensor design technology developed in the Baker lab, was led by Baker lab postdoctoral scholars Jason Zhang, PhD, and Hsien-Wei (Andy) Yeh, PhD. From Behind…
-
Rotory proteins designed from scratch
Today we report in Science the design of rotary devices made from custom proteins. These microscopic “axles” and “rotors” come together to form spinning assemblies, rather than being locked in just one orientation. Such mechanical coupling is a key feature of any machine. The new axle-rotor devices — which are…
-
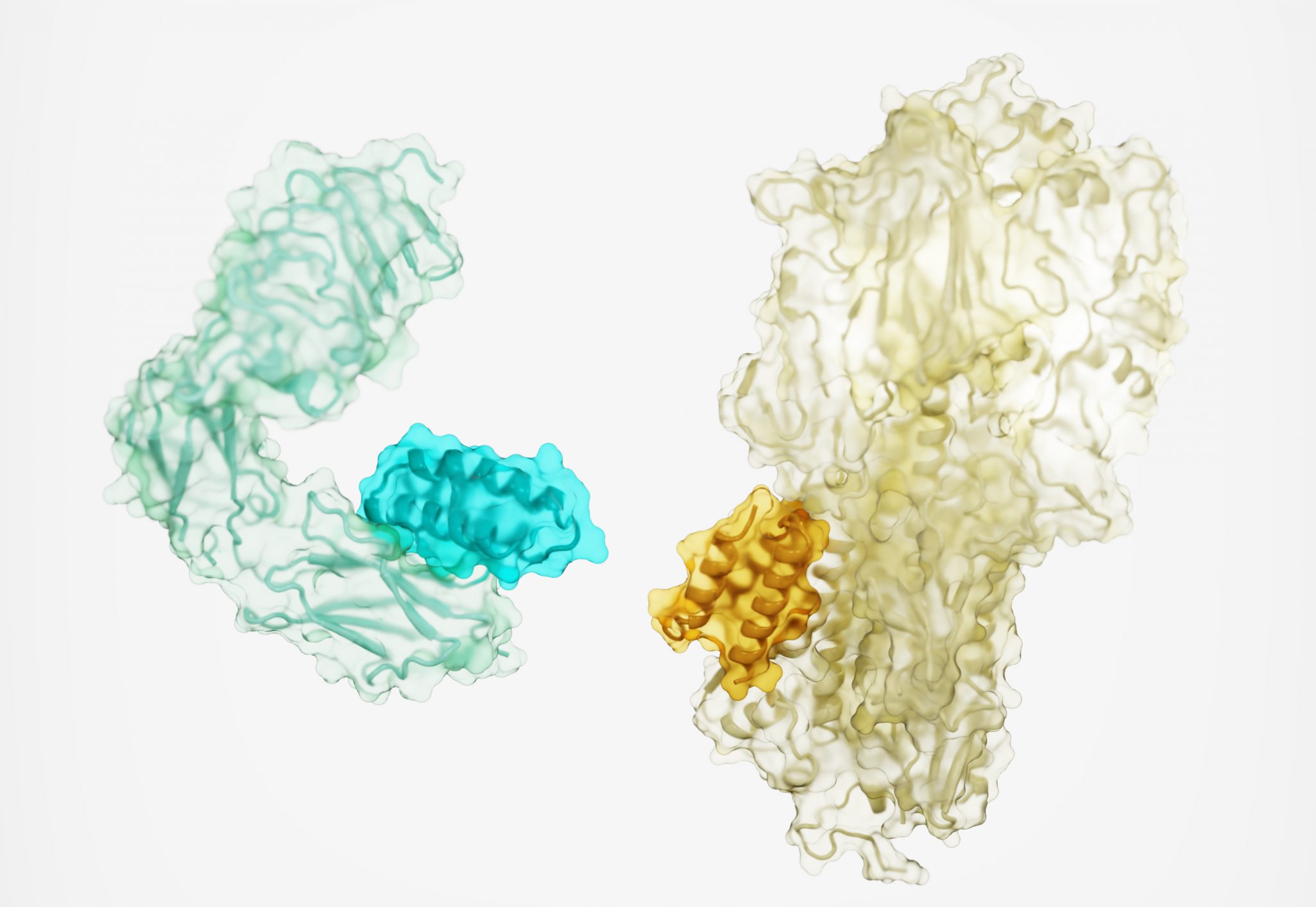
Protein drugs designed from the ground up
Today we report in Nature a new method for generating protein drugs. Using Rosetta-based design, an international team designed molecules that can target important proteins in the body, such as the insulin receptor, as well as proteins on the surface of viruses. This solves a long-standing challenge in drug development and may…
-
Diverse protein assemblies by (negative) design
A new approach for creating custom protein complexes yields asymmetric assemblies with interchangeable parts. Today we report in Science the design of new protein assemblies made from modular parts. These complexes — which adopt linear, branching, or closed-loop architectures — contain up to six unique proteins, each of which remains folded…
-
Breakthrough of the Year
The journal Science has selected artificial intelligence algorithms that predict the three-dimensional shapes of proteins — as well as the blizzard of protein structures they have revealed — as their 2021 Breakthrough of the Year. We are honored to have our work in this field recognized alongside that of the…
-
Deep learning dreams up new protein structures
Just as convincing images of cats can be created using artificial intelligence, new proteins can now be made using similar tools. In a new report in Nature, we describe the development of a neural network that “hallucinates” proteins with new, stable structures. “For this project, we made up completely random…
-
UW BIOFAB: a force for reproducible science
This article was written by Renske Dyedov (UW) Key to advancing any new scientific discovery is the ability for researchers to independently repeat the experiments that led to it. In science today, particularly biology, the lack of reproducibility between experiments is a major problem that slows scientific progress, wastes resources…
-
Deep learning reveals how proteins interact
A team led by scientsts in the Baker lab has combined recent advances in evolutionary analysis and deep learning to build three-dimensional models of how most proteins in eukaryotes interact. This breakthrough has significant implications for understanding the biochemical processes that are common to all animals, plants, and fungi. This…