Author: admin
-
Accurate Design of Co-Assembling Multi-Component Protein Nanomaterials
A new paper is out in the June 5 issue of Nature entitled Accurate design of co-assembling multi-component protein nanomaterials. Scientists at the Institute for Protein Design (IPD), in collaboration with researchers at UCLA and HHMI, have built upon their previous work constructing single-component protein nanocages and can now design and build self-assembling protein nanomaterials made up of…
-
May IPD News Roundup
May was a busy month at the IPD, with some new publications and exciting announcements! Learn more here.
-
Removing T-cell Epitopes with Computational Protein Design
In a recent PNAS paper entitled “Removing T-cell epitopes with computational protein design”, IPD researchers combine machine learning with computational protein design to demonstrate immune silencing of protein targets. This deimmunization has the potential to reduce or eliminate immunogenicity of protein therapeutics. Learn more at this link.
-
Women in Science Lunch Discussion
A recent Nature issue exposed the dismaying fact that many women are deterred from pursuing a career in science, especially at the highest levels (postdoctoral positions, faculty position, scientific advisory boards to start up companies, etc). To talk about this significant gender gap in science and the issues female scientists face, Baker lab members participated…
-
WRF Awards $8M for the IPD Innovation Fellows Program
May 15, 2014 With a very generous $8 M gift from the Washington Research Foundation (WRF), the IPD has launched the WRF-IPD Innovation Fellows Program supporting research partnerships between the IPD and other Seattle-area research institutes or UW departments. We are recruiting exceptionally talented researchers who have just finished their PhD to join expert laboratories at local institutions where they…
-
The “Three Dreamer” Protein Design Partnership for Alzheimer’s Disease
The “Three Dreamers” are a group of Seattle-based philanthropists whose family members are suffering from Alzheimer’s disease (AD). The IPD has partnered with the Three Dreamers, the Foldit community and AD researchers at the UW to design new proteins targeting amyloid, thought to be the cause of AD. Learn more at this link.
-
Beyond Evolution: Protein Design News and Art
Re/Code writer James Temple has written an interesting article on David Baker’s efforts to design a new world of proteins. The article covers the IPD efforts to design proteins that neutralize the flu virus, Alzheimer’s disease amyloid protein, and how the IPD is engaging citizen scientists in the Rosetta@home and Foldit projects. Learn more at…
-
Design of Activated Serine-Containing Catalytic Triads with Atomic-Level Accuracy
Baker lab members published in Nature Chemical Biology a paper entitled “Design of activated serine-containing catalytic triads with atomic-level accuracy“, describing the computational design of proteins with idealized serine-containing catalytic triads which can capture and neutralize organophosphate probes. This work has utility in design of scavengers of environmental toxins. Learn more at this link.
-
Increasing Public Involvement in Structural Biology
Foldit is 5 years old. This publication entitled “Increasing public involvement in structural biology” chronicles the power of engaging the citizen science community on behalf of the computational challenge of protein folding. Learn more at this link.
-
A New Vaccine Design Method To Combat A Dangerous Virus
In a widely cited Nature paper entitled Proof of principle for epitope-focused vaccine design, IPD researchers and collaborators invented a new method to design novel proteins for use as a candidate vaccines to protect against respiratory syncytial virus (RSV), a significant cause of infant mortality. Learn more at this link.
-
Seattle Health Innovators Visit the IPD
The Seattle Health Innovators recently visited the Institute for Protein Design, and wrote a nice blog entitled “Crowdsourcing the design of new molecules to improve healthcare and the environment.” The article provides a nice look into the Institute for Protein Design. Learn more at this link.
-
Computational Design of a pH Sensitive Antibody Binder
Purification of antibody IgG from crude serum or culture medium is required for virtually all research, diagnostic, and therapeutic antibody applications. Researchers at the Institute for Protein Design (IPD) have used computational methods to design a new protein (called “Fc-Binder”) that is programed to bind to the constant portion of IgG (aka “Fc” region) at basic…
-
KumaMax: Winner of C4C Recognition as a Novel Oral Therapeutic for Celiac Disease
Dr. Ingrid Swanson Pultz, a Translational Investigator at the Institute for Protein Design won first prize at the UW Center for Commercialization 2013 Innovator Recognition Event, for KumaMax, an enzyme designed in the Baker lab to efficiently break down gluten within the acidic environment of the stomach, before it can reach the small intestine where intact gluten…
-
David Baker at the 2013 Gairdner Award Celebrations in Toronto
David Baker, Head of the Institute for Protein Design was recently in Toronto, Canada in late October to deliver a lecture on protein design as part of Gairdner Award celebrations. This was written up in the Globe and Mail. Learn more at this link.
-
GIVING IN ACTION: Making Dreams Come True at the IPD
Thank you to our supporters. GIVING IN ACTION: Making Dreams Come True. Barton Family Foundation, Life Sciences Discovery Fund, Bruce and Jeannie Nordstrom, and Three Dreamers all supporting the IPD. Thank you all for your help in supporting our efforts in protein design.
-
Computational Protein Design To Improve Detoxification Rates Of Nerve Agents
V-type nerve agents are among the most toxic compounds known, and are chemically related to pesticides widespread in the environment. Using an integrated approach, described in an ACS Chemical Biology paper entitled Engineering V-type nerve agents detoxifying enzymes using computationally focused libraries, Dr. Izhack Cherny, Dr. Per Greisen, and collaborators increased the rate of nerve…
-
Automating Human Intuition for Protein Design
IPD researchers in the Baker group have improved protein computational design algorithms by “Automating human intuition for protein design.” Read more at this link.
-
Follow the Institute for Protein Design on Facebook and Twitter
Visit us on Facebook and follow us on Twitter @UWproteindesign
-
Professor David Baker Provides an In Depth Discussion on Protein Design
Prof. David Baker, Head of the UW Institute for Protein Design, HHMI Investigator provides an in depth discussion on the design of protein structures, functions and assemblies. Click here to watch the video.
-
Institute for Protein Design Infograph
[soliloquy id=”345″]
-
High-Resolution Comparative Modeling with RosettaCM
Researchers in the Baker group describe an improved method for comparative modeling, RosettaCM, which optimizes a physically realistic all-atom energy function over the conformational space defined by homologous structures. Learn more at this link.
-
Computational Design Of A Protein That Binds Polar Surfaces
In a Journal of Molecular Biology publication entitled Computational design of a protein-based enzyme inhibitor, Dr. Erik Procko and collaborators describe the computational design of a protein-based enzyme inhibitor that binds the polar active site of hen egg lysosome (HEL). Computational design of a protein that binds polar surfaces has not been previously accomplished. Learn…
-
Rosetta Designed Protein (Toca 511), Now Offering Hope to Brain Cancer Patients
Brain cancer is a serious unmet medical challenge, and Washington state is one of the leading research clusters working on glioblastoma. Here we report on how RosettaDesign proteins are being used to treat brain cancer! Read more about this important translational protein design effort here.
-
Life Sciences Discovery Fund Awards $1.4M to the Institute for Protein Design
The Life Sciences Discovery Fund (LSDF) today announced its latest round of Opportunity Grants, and awarded $1.4 M to the University of Washington (UW) to “Launch of the Institute for Protein Design for Creating New Therapeutics, Vaccines and Diagnostics.” This LSDF Opportunity Grant Award will enable the IPD Translational Investigators to improve upon protein design discoveries so that they…
-
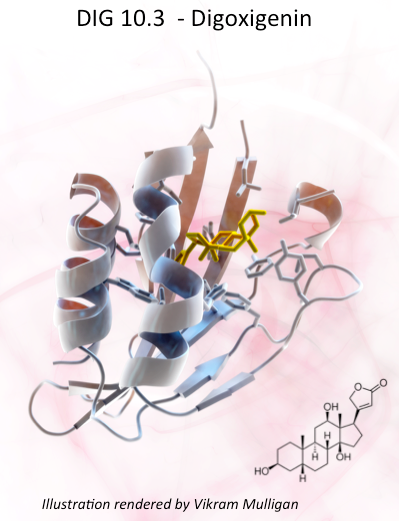
One Small Molecule Binding Protein, One Giant Leap for Protein Design
Reported on-line in Nature (Sept. 4, 2013) researchers at the Institute for Protein Design describe the use of Rosetta computer algorithms to design a protein which binds with high affinity and specificity to a small drug molecule, digoxigenin a dangerous but sometimes life saving cardiac glycoside. Learn more at this link.
-
IPD Researchers Publish New Protocols for Preparing Protein Scaffold Libraries for Functional Site Design
IPD researchers in the Baker group have published new computational protocols for preparing protein scaffold libraries for functional site design. Their paper entitled “A Pareto-optimal refinement method for protein design scaffolds“ improves the search for amino acids with the lowest energy subject to a set of constraints specifying function. Learn more at this link.
-
Centenary Award and Frederick Gowland Hopkins Memorial Lecture
Dr. David Baker, Director of the IPD delivered the Centenary Award and Frederick Gowland Hopkins Memorial Lecture at at the MRC Laboratory of Molecular Biology, Cambridge, UK, on December, 13, 2012. Baker’s lecture entitled “Protein folding, structure prediction and design” can be read at this published link. See: Baker, D. (2014). Protein folding, structure prediction and design.. Biochemical Society…
-
Proteins Made to Order. Researchers at the IPD Design Proteins from Scratch with Predictable Structures
A team from David Baker’s laboratory at the University of Washington in Seattle have described a set of “rules” for the design of proteins from scratch, and have demonstrated the successful design of five new proteins that fold reliably into predicted conformations. Their work was published Nature. Learn more at this link.
-

The IPD Moves Into the New Molecular Engineering and Sciences Building
The Institute for Protein Design has moved into the new Molecular Engineering & Sciences Building located in the heart of the University of Washington Seattle campus.
-
Computer Designed Proteins Programmed to Disarm a Variety of Flu Viruses
As reported in Nature Biotechnology, David Baker and scientists at the IPD published exciting new methods to improve the potency and breadth of computer-designed protein inhibitors of influenza. Learn more at this link.
-
Computational Design of Self-Assembling Protein Nanomaterials with Atomic Level Accuracy
IPD researchers in the Baker group have published in Science a paper entitled “Computational design of self-assembling protein nanomaterials with atomic level accuracy.” They describe a general computational method for designing proteins that self-assemble to a desired symmetric architecture. Protein building blocks are docked together symmetrically to identify complementary packing arrangements, and low-energy protein-protein interfaces are then designed…
-
UW establishes the Institute for Protein Design
Hello, world.
