Author: admin
-
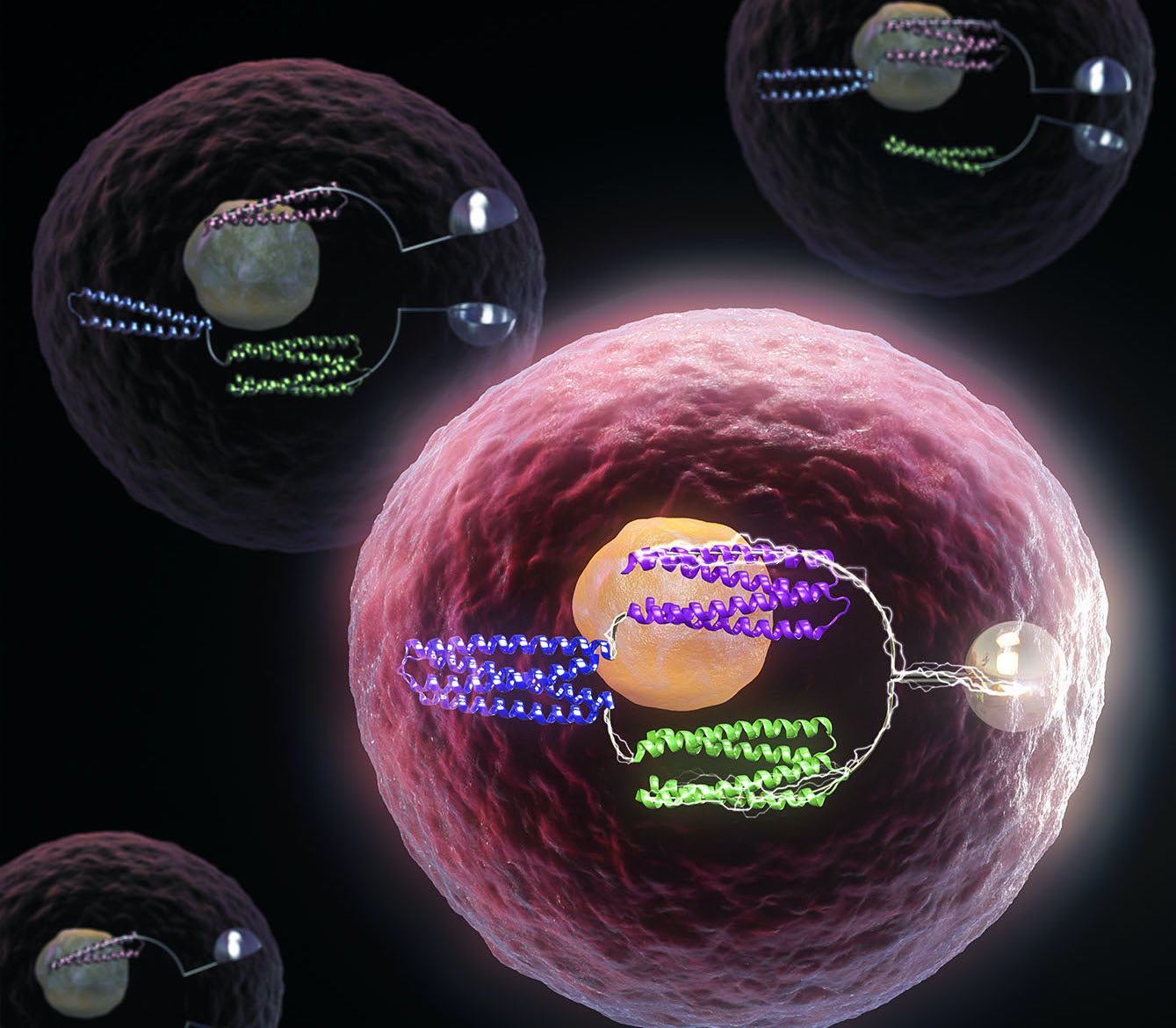
De novo design of protein logic gates
The same basic tools that allow computers to function are now being used to control life at the molecular level, with implications for future medicines and synthetic biology. Together with collaborators, we have created artificial proteins that function as molecular logic gates. These tools, like their electronic counterparts, can be used to program the behavior…
-

Volunteers rally to Rosetta@Home to stop COVID-19
As schools, museums, offices and stores shutter to slow the spread of the new coronavirus, millions of people are now finding themselves stuck at home. Fortunately, even in these trying times, there are are small steps that anyone can be take to help combat COVID-19. One option is to donate to biomedical research — but…
-
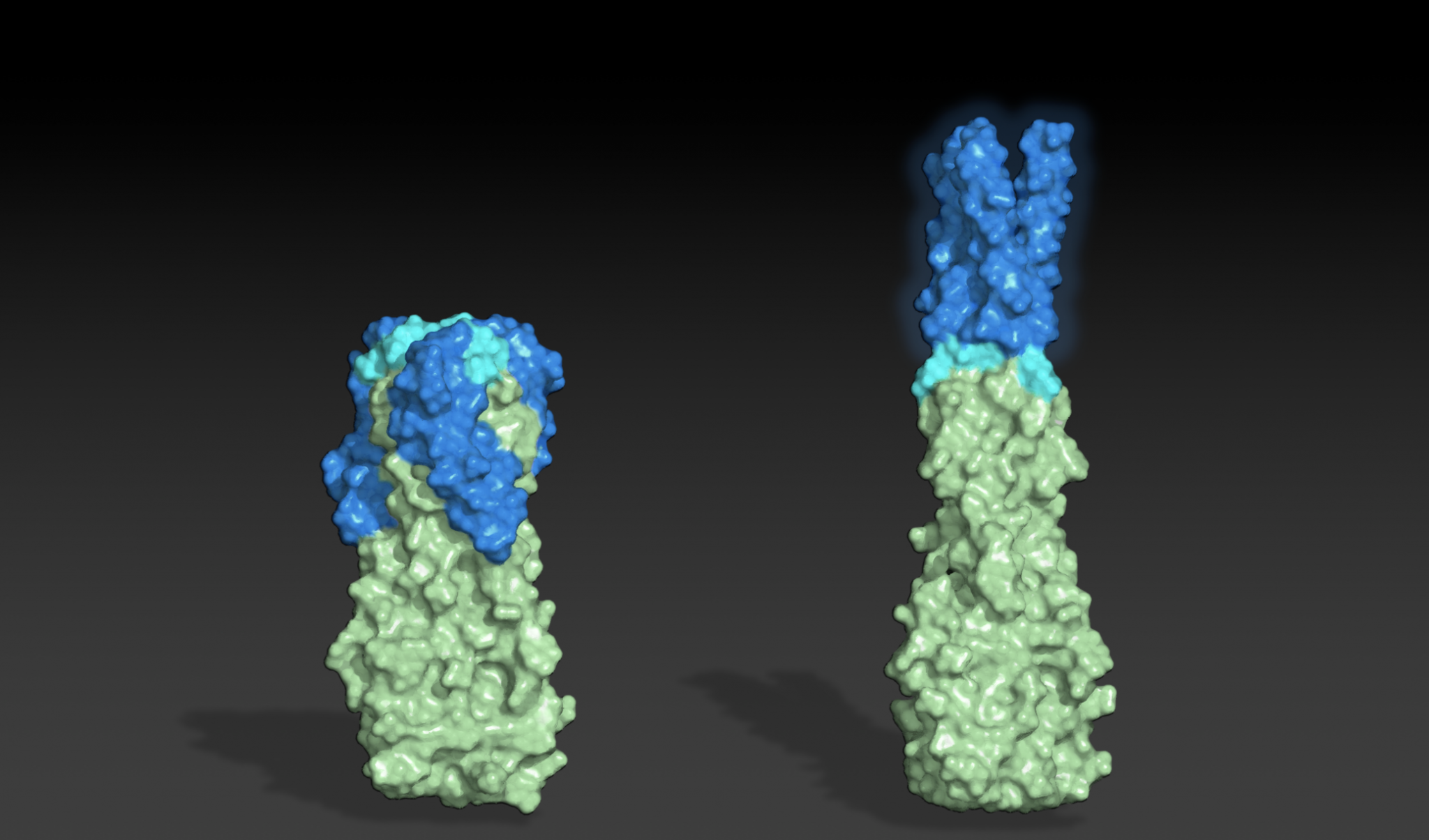
Designing shape-shifting proteins
Today we report the design of protein sequences that adopt more than one well-folded structure, reminiscent of viral fusion proteins. This research moves us closer to creating artificial protein systems with reliable moving parts. In nature, many proteins change shape in response to their environment. This plasticity is often linked to biological function. While computational…
-

Play Foldit to help stop coronavirus (VIDEO)
You don’t have to be a scientist to do science! By playing the computer game Foldit, you can help discover new antiviral drugs that might stop the novel coronavirus. The most promising solutions will be manufactured and tested at the University of Washington Institute for Protein Design in Seattle. Foldit is run by academic research scientists. It is free…
-
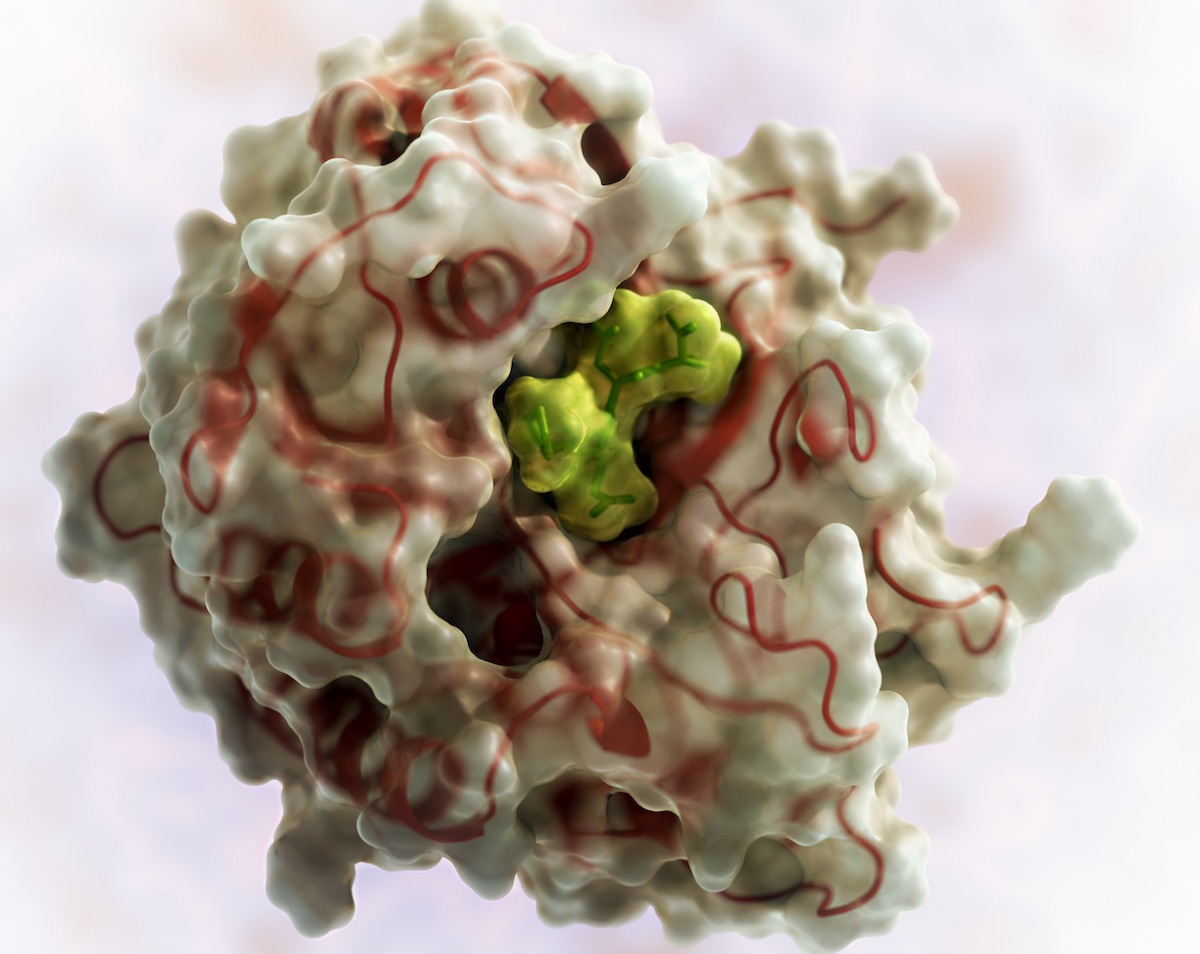
Takeda acquires PvP Biologics
PvP Biologics is on a mission to develop a highly-effective therapeutic to reduce the burden of living with celiac disease. They are advancing an oral enzyme — TAK-062 — designed to break down gluten in the stomach. This exciting research, which began as an iGEM project in 2011, was matured at the Institute for Protein…
-
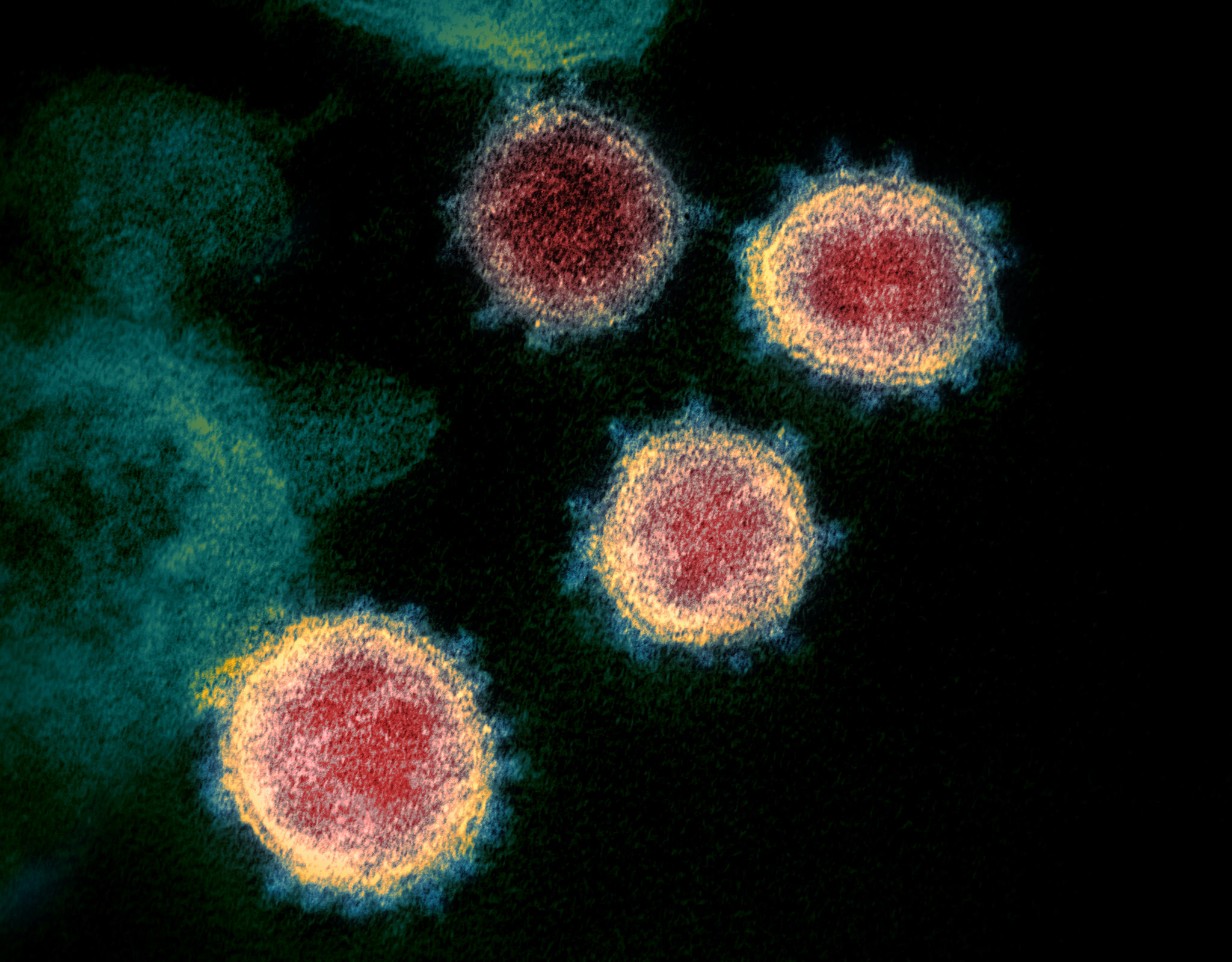
Rosetta’s role in fighting coronavirus
We are happy to report that the Rosetta molecular modeling suite was recently used to accurately predict the atomic-scale structure of an important coronavirus protein weeks before it could be measured in the lab. Knowledge gained from studying this viral protein is now being used to guide the design of novel vaccines and antiviral drugs.…
-

Introducing WE-REACH, a new biomedical innovation hub
With $4 million in matching funds from the National Institutes of Health, the University of Washington has created a new integrated center to match biomedical discoveries with the resources needed to bring innovative products to the public and improve health. “The University of Washington and regional partner institutions produce some of the most exciting biomedical…
-

SCI-STEM Symposium 2020
The Institute for Protein Design at the University of Washington held the second symposium aimed at providing strategies to address diversity challenges in science, technology, engineering, and math (STEM). The Strategies for Cultivating Inclusion in STEM (SCI-STEM) symposium featured leading keynote speakers, panel discussions, and interactive breakout sessions. Members of the STEM community at all…
-
Radhika wins poster award at ABRCMS
Undergraduate researcher Radhika Dalal took home a poster award at this year’s Annual Biomedical Research Conference for Minority Students in Anaheim, California. Her mentors include IPD graduate student Una Natterman and Quinton Dowling. Way to go, Radhika! And congratulations to all the award winners:
-
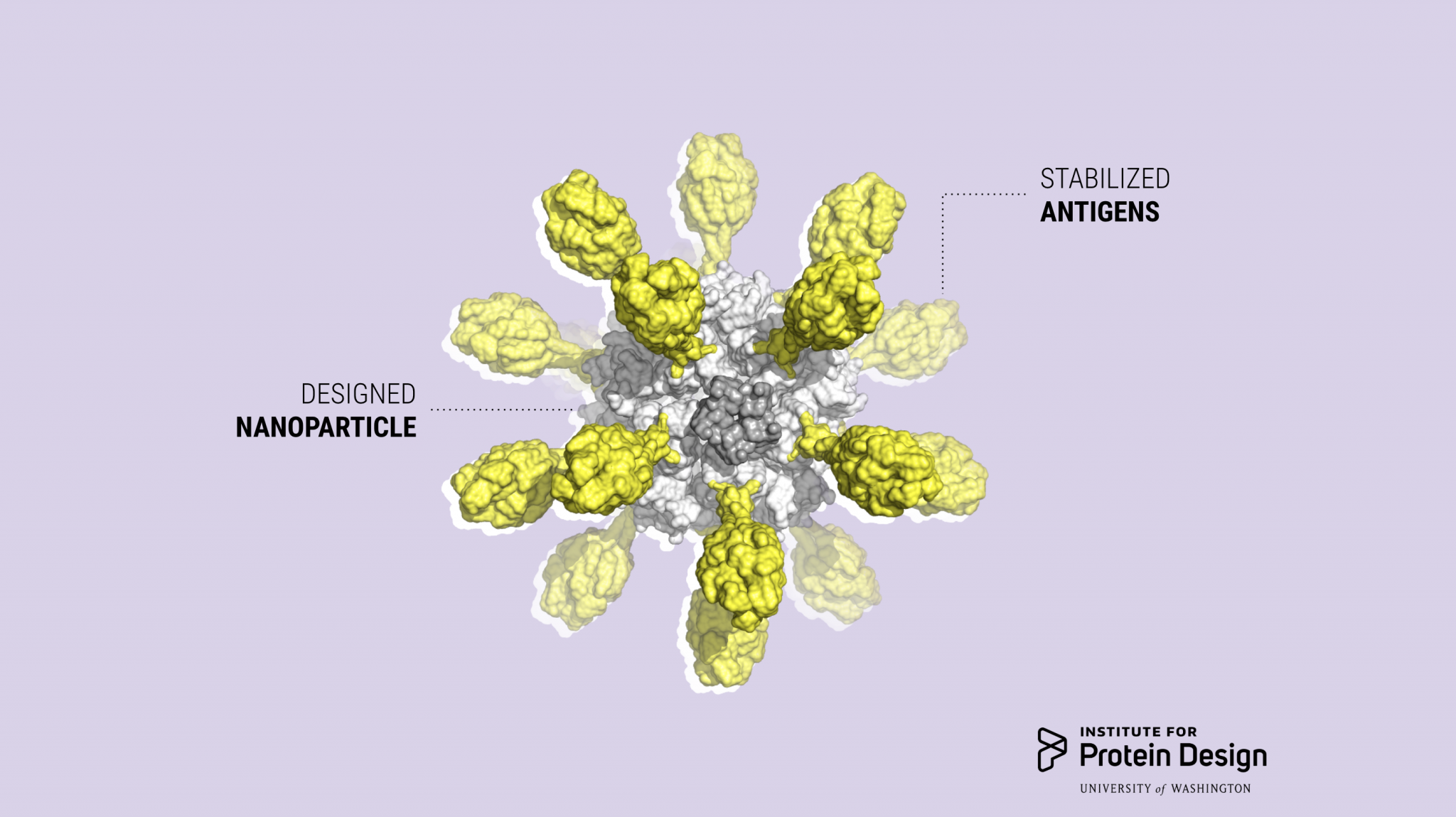
Icosavax launches to advance designer vaccines
Icosavax, Inc. today announced its launch with a $51 million Series A financing. The company was founded on computationally designed self-assembling virus-like particle (VLP) technology developed here at the IPD (Cell 2019, Preview). The proceeds of the financing will be used to advance the company’s first vaccine candidate, IVX-121, for respiratory syncytial virus (RSV) for…
-
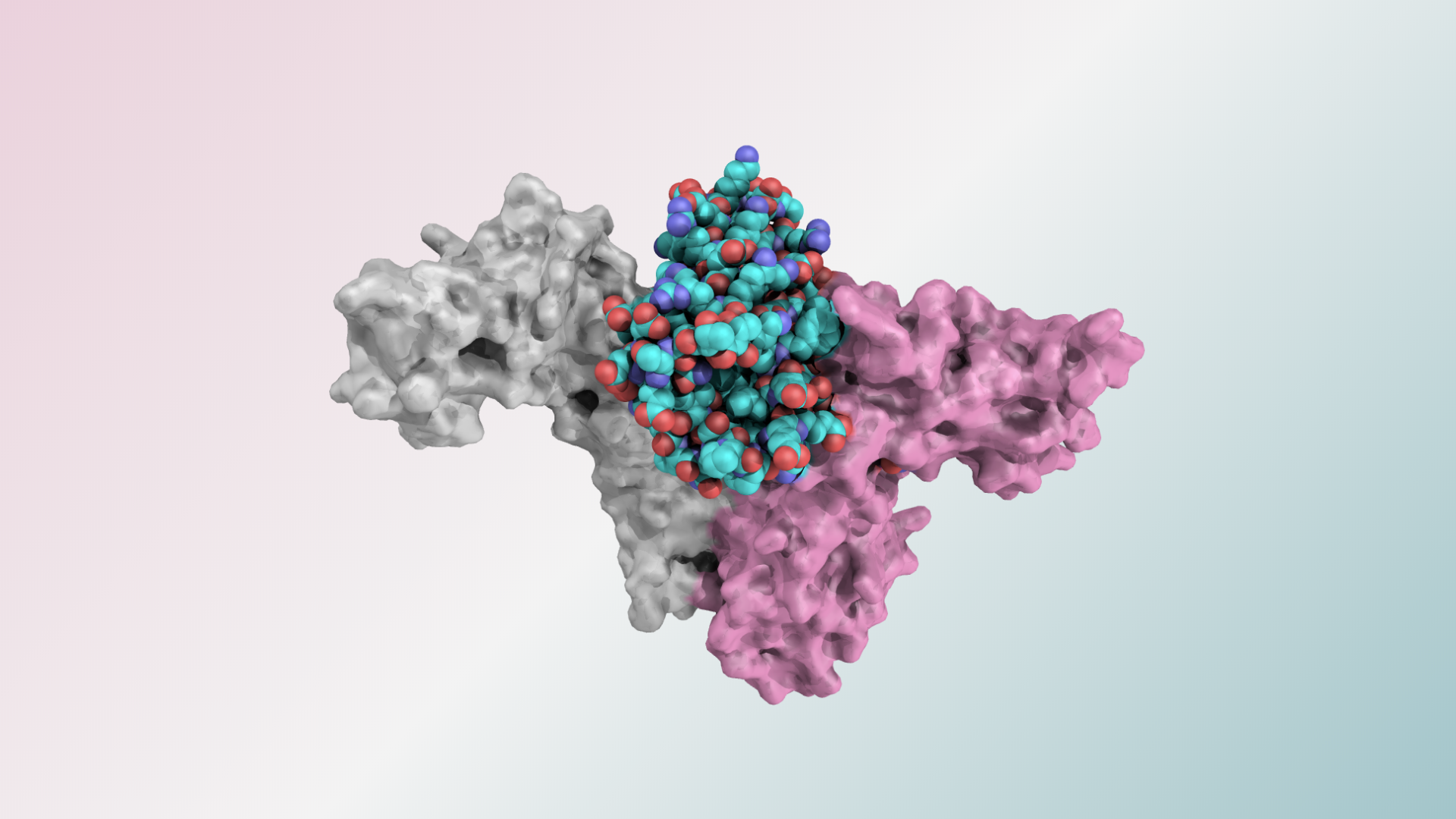
Neoleukin: from spinout to public company in 7 months
IPD-spinout Neoleukin Therapeutics announced this week a merger with Aquinox Pharmaceuticals, a publicly traded company. The combined company will change its name to Neoleukin Therapeutics, and will continue to advance its Rosetta-designed protein platform for cancer, inflammation, and autoimmune diseases. Neoleukin was spun out of the IPD Translational Investigator Program in January. As a result…
-

5 questions about LOCKR from our Reddit AMA
Researchers from the IPD and UCSF recently participated in a Reddit Ask Me Anything about LOCKR, our new de novo protein switch. Reddit users had dozens of fantastic questions — so many, in fact, that the team ran out of time before they could address them all. “The questions were both insightful and interesting,” says…
-

Introducing LOCKR: a bioactive protein switch
Today we report in Nature the design and initial applications of the first completely artificial protein switch that can work inside living cells to modify—or even commandeer—the cell’s complex internal circuitry. The switch is dubbed LOCKR, short for Latching, Orthogonal Cage/Key pRotein. “In the same way that integrated circuits enabled the explosion of the computer chip industry, these versatile and dynamic…
-

Feature: The computational protein designers
Jeffrey Perkel, technology editor at Nature, has just written a feature on de novo protein design. PDF
-
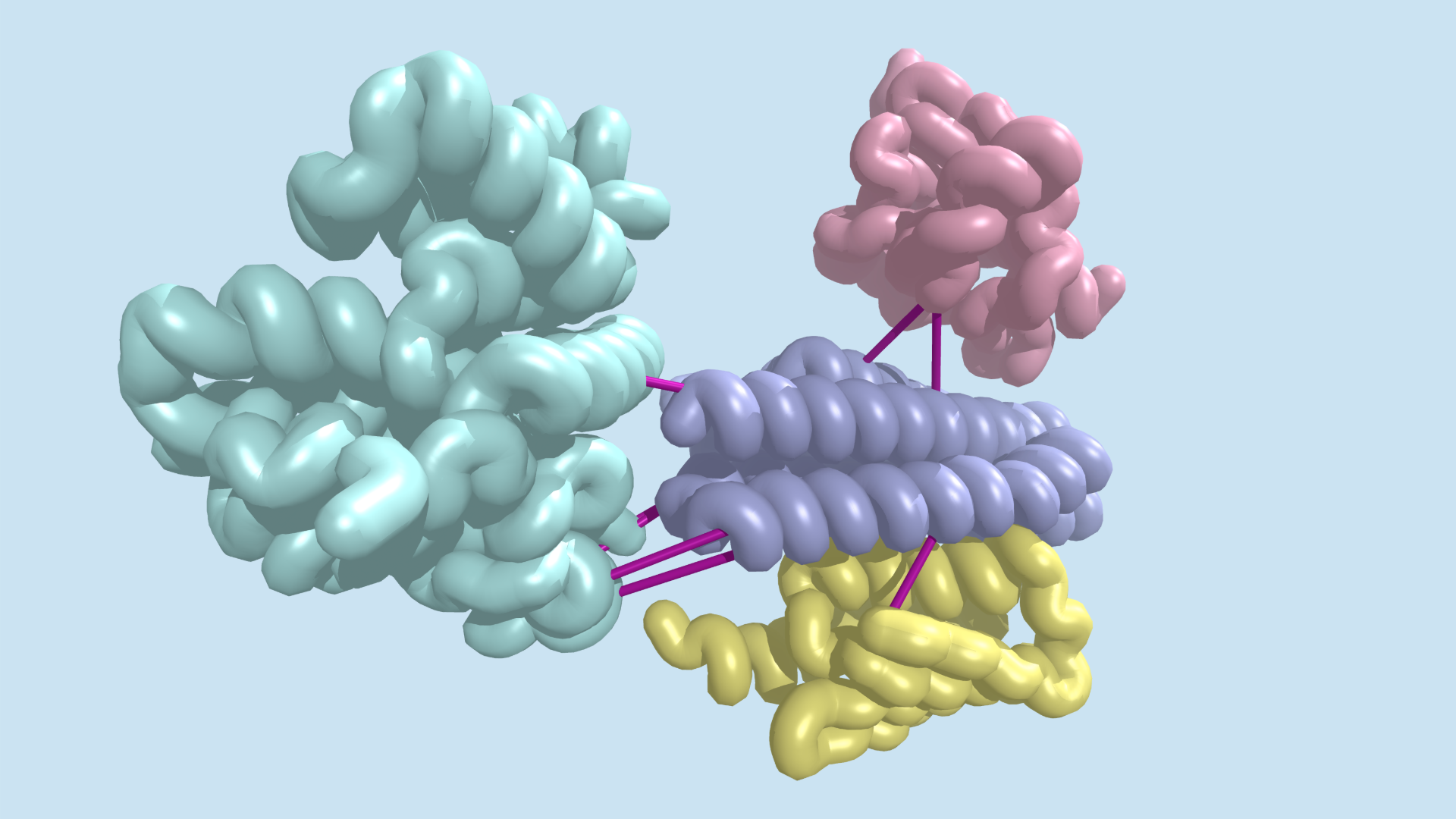
Coevolution at the proteome scale
Today we report in Science the identification of hundreds of previously uncharacterized protein–protein interactions in E. coli and the pathogenic bacterium M. tuberculosis. These include both previously unknown protein complexes and previously uncharacterized components of known complexes. This research was led by postdoctoral fellow Qian Cong and included former Baker lab graduate student Sergey Ovchinnikov, now a John Harvard Distinguished Science…
-

Protein arrays on mineral surfaces
Today we report the design of synthetic protein arrays that assemble on the surface of mica, a common and exceptionally smooth crystalline mineral. This work, which was performed in collaboration with the De Yoreo lab at PNNL, provides a foundation for understanding how protein-crystal interactions can be systematically programmed. Our goal was to engineer artificial proteins to self-assemble on…
-
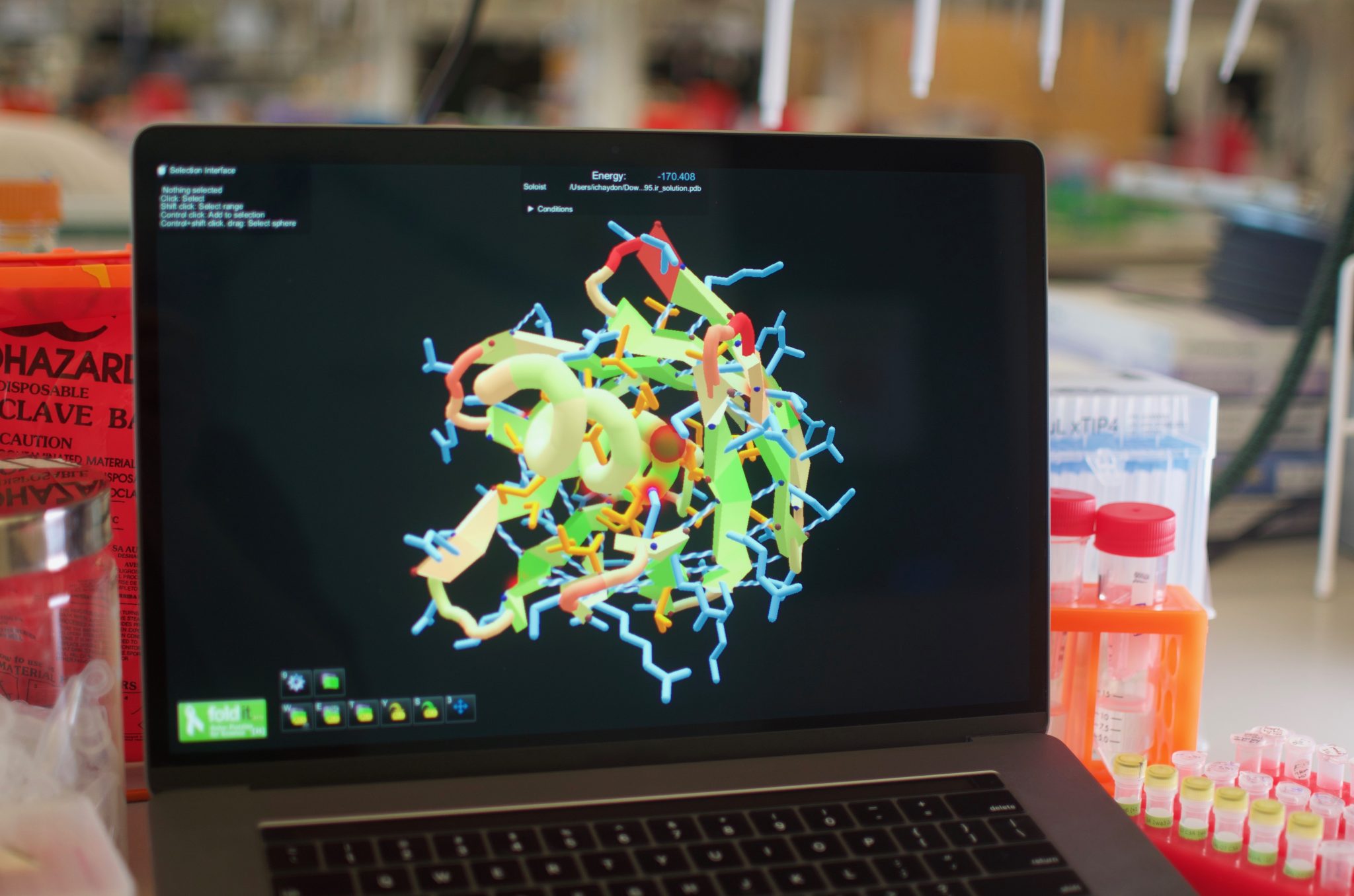
Protein design by citizen scientists
Citizen scientists can now use Foldit to successfully design synthetic proteins. The initial results of this unique collaboration appear today in Nature. Brian Koepnick, a recent PhD graduate in the Baker lab, led a team that worked on Foldit behind the scenes, introducing new features into the game that they believed would help players home in on…
-
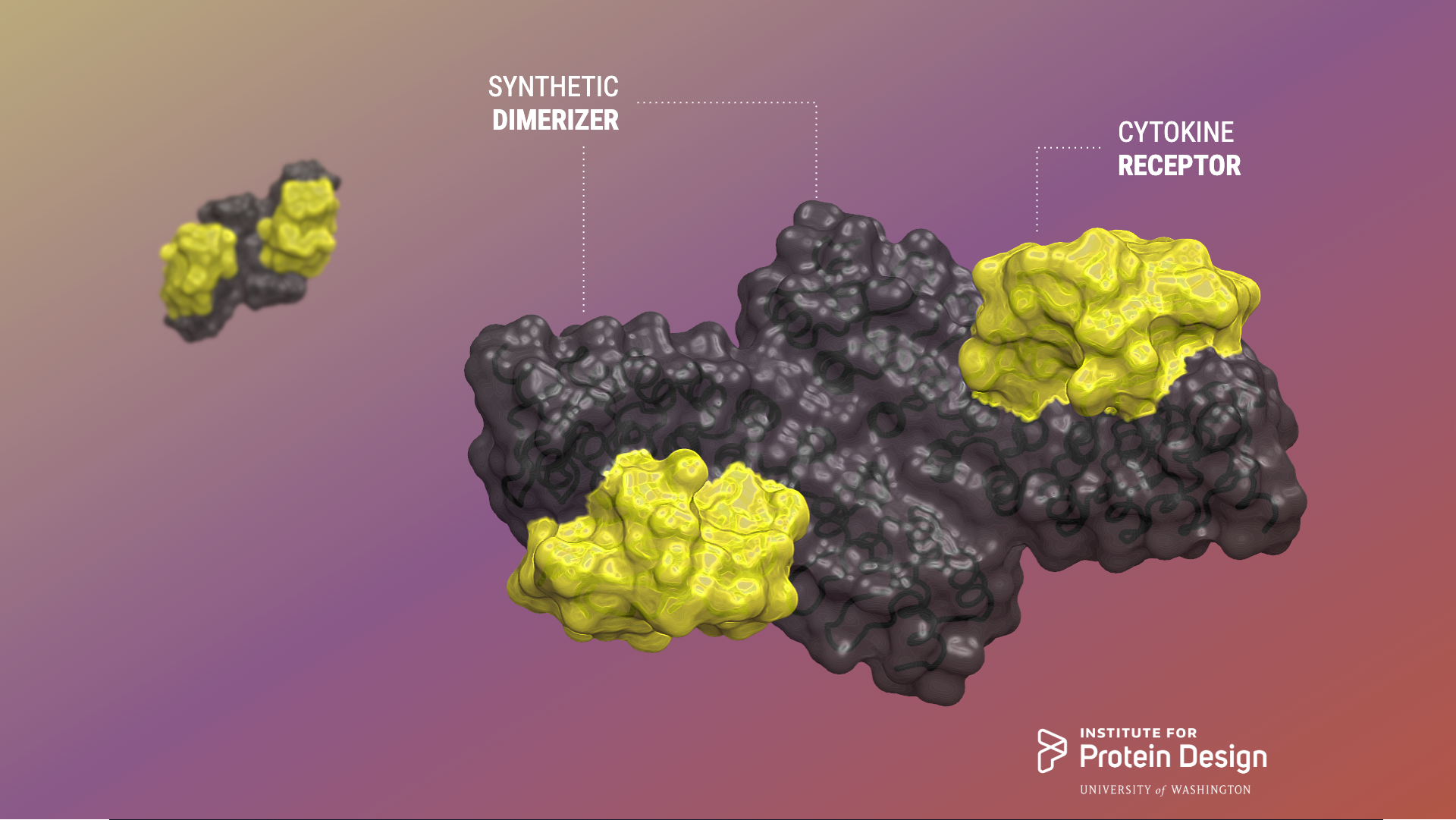
Designed ligands tune cytokine signaling
Today the Baker lab shares some exciting collaborative results of their efforts to design rigid and tunable receptor dimerizers. The first authors of this report are Kritika Mohan, Stanford, and George Ueda, IPD. From Science: Exploring a range of signaling Cytokines are small proteins that bind to the extracellular domains of transmembrane receptors to activate signaling pathways…
-
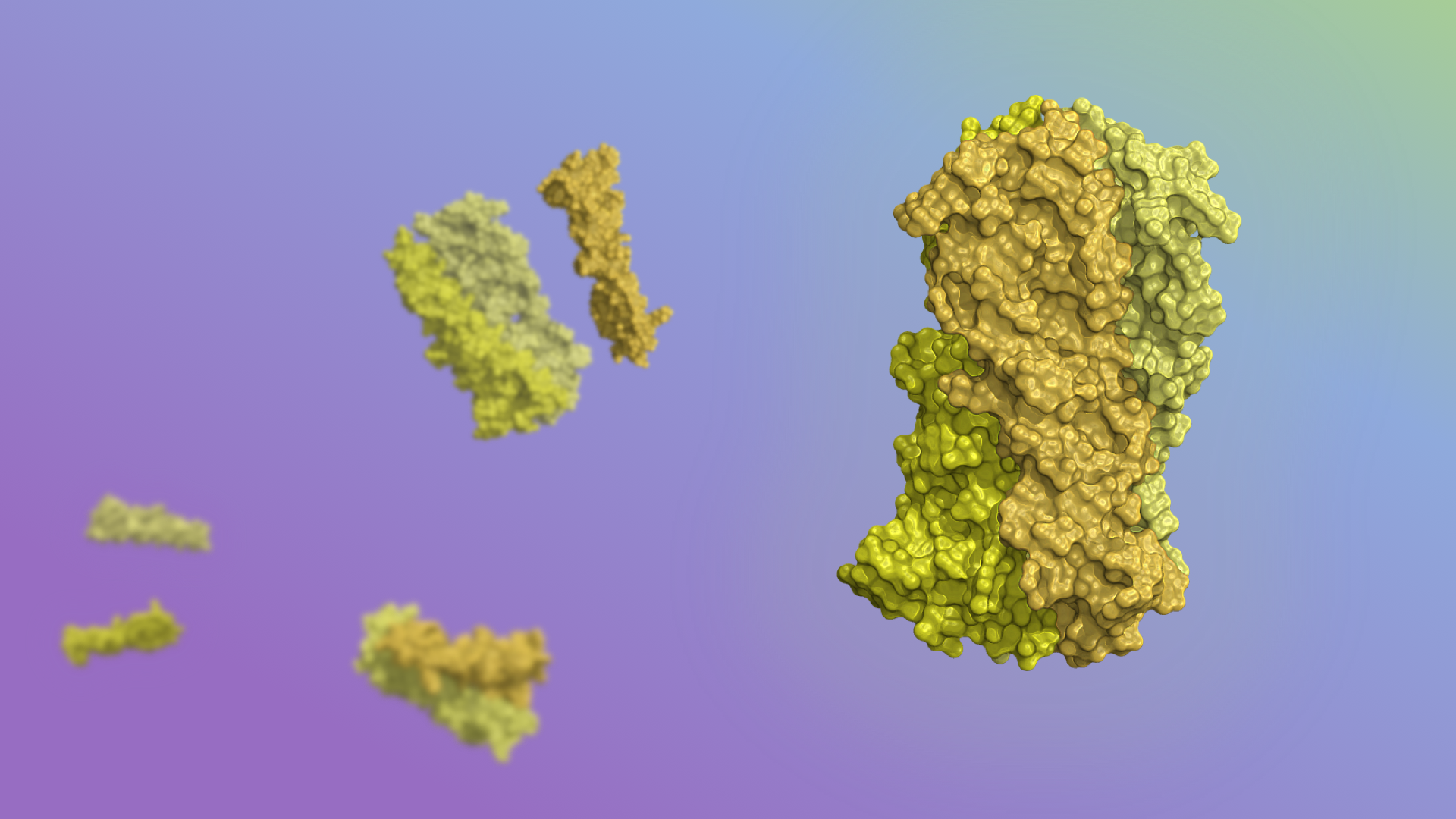
Tunable pH-dependent assemblies
Natural proteins often shift their shapes in precise ways in order to function. Achieving similar molecular rearrangements by design, however, has been a long-standing challenge. Today, a team of researchers lead by scientists at the IPD report in Science the rational design of synthetic proteins that move in response to their environment in predictable and tunable…
-
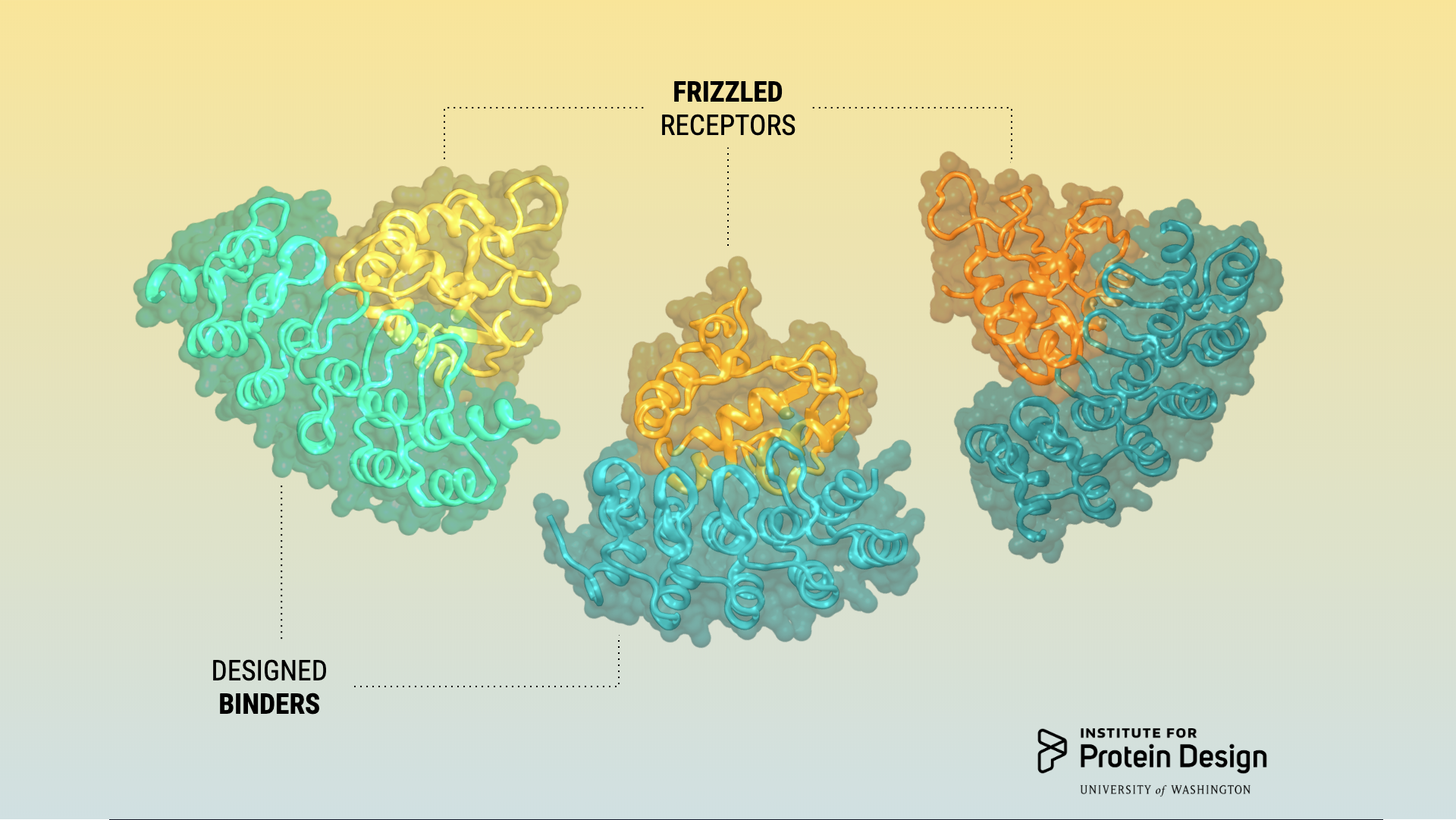
Receptor sub-type binders
This week we report in NSMB a combined computational design and experimental selection approach for creating proteins that bind selectively to closely related receptor subtypes. This project was led by Luke Dang, a former Baker lab graduate student, and Yi Miao, a postdoctoral researcher in Christopher Garcia’s lab at Stanford. Abstract: To discriminate between closely related members of…
-
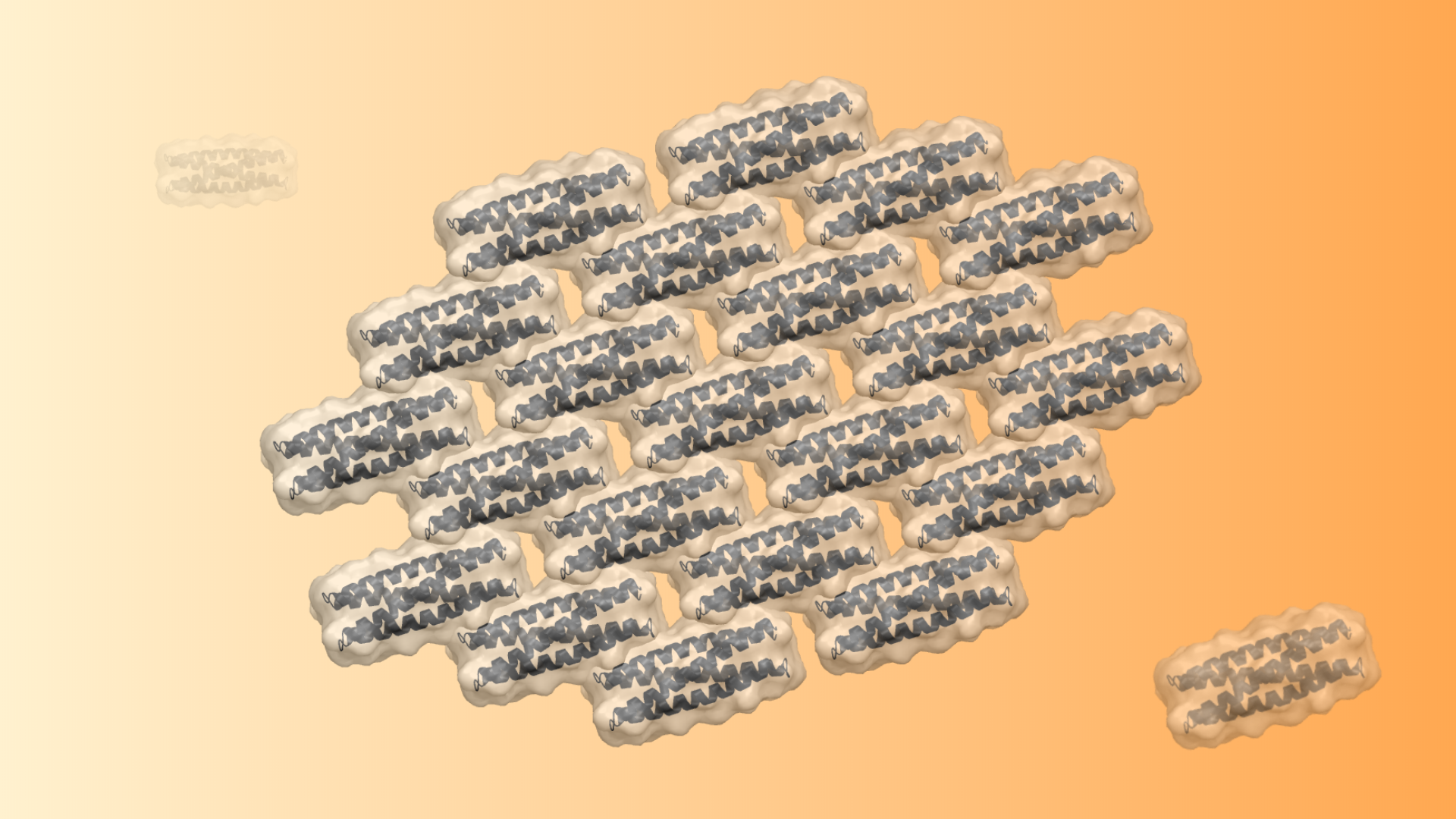
De novo 2D arrays
This week we report in JACS a general approach for designing self-assembling 2D protein arrays. This project was led by Zibo Chen, a recent Baker lab graduate student, and featured collaborators from the, DiMaio, De Yoreo and Kollman labs at UW. Abstract: Modular self-assembly of biomolecules in two dimensions (2D) is straightforward with DNA but…
-

Our outstanding postdoc mentors
We’re thrilled to share that four members of our Institute have been nominated for the UW Graduate School’s Postdoc Mentoring Award. Each brings invaluable guidance and advice to their graduate student and undergraduate trainees. This Year’s Winner: Gabriella Wolff, Biology Finalists: Michael Beyeler, Psychology David Grossnickle, Biology Matthew Hart, Pathology Karla-Luise Herpoldt, Biochemistry Kelly Hines,…
-

Introducing our Audacious Project
We’ve been selected to join The Audacious Project, a philanthropic collaborative organized by TED. Read all about our project here. In short, we’re expanding our institute into a global hub of innovation so that protein design can be applied to help solve some of the world’s most pressing challenges. Our director David Baker gave an…
-
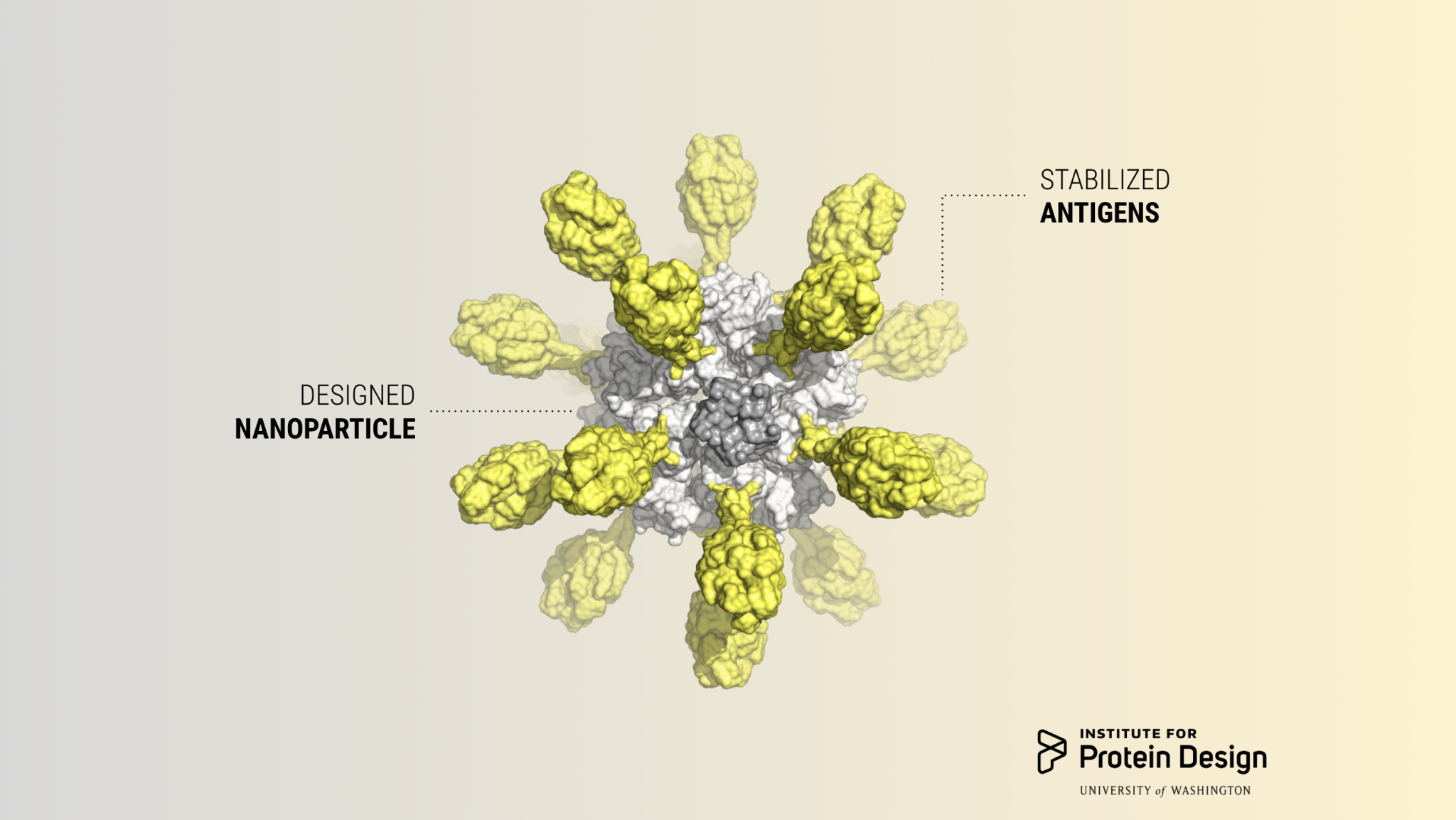
Designing a stable and potent RSV vaccine candidate
Today we report in Cell our first computer-designed nanoparticle vaccine targeting respiratory syncytial virus, the primary cause of pneumonia in young children and the leading cause of infant mortality worldwide after malaria. Although virtually every child will get infected by RSV before the age of three, an estimated 99 percent of deaths associated with the…
-
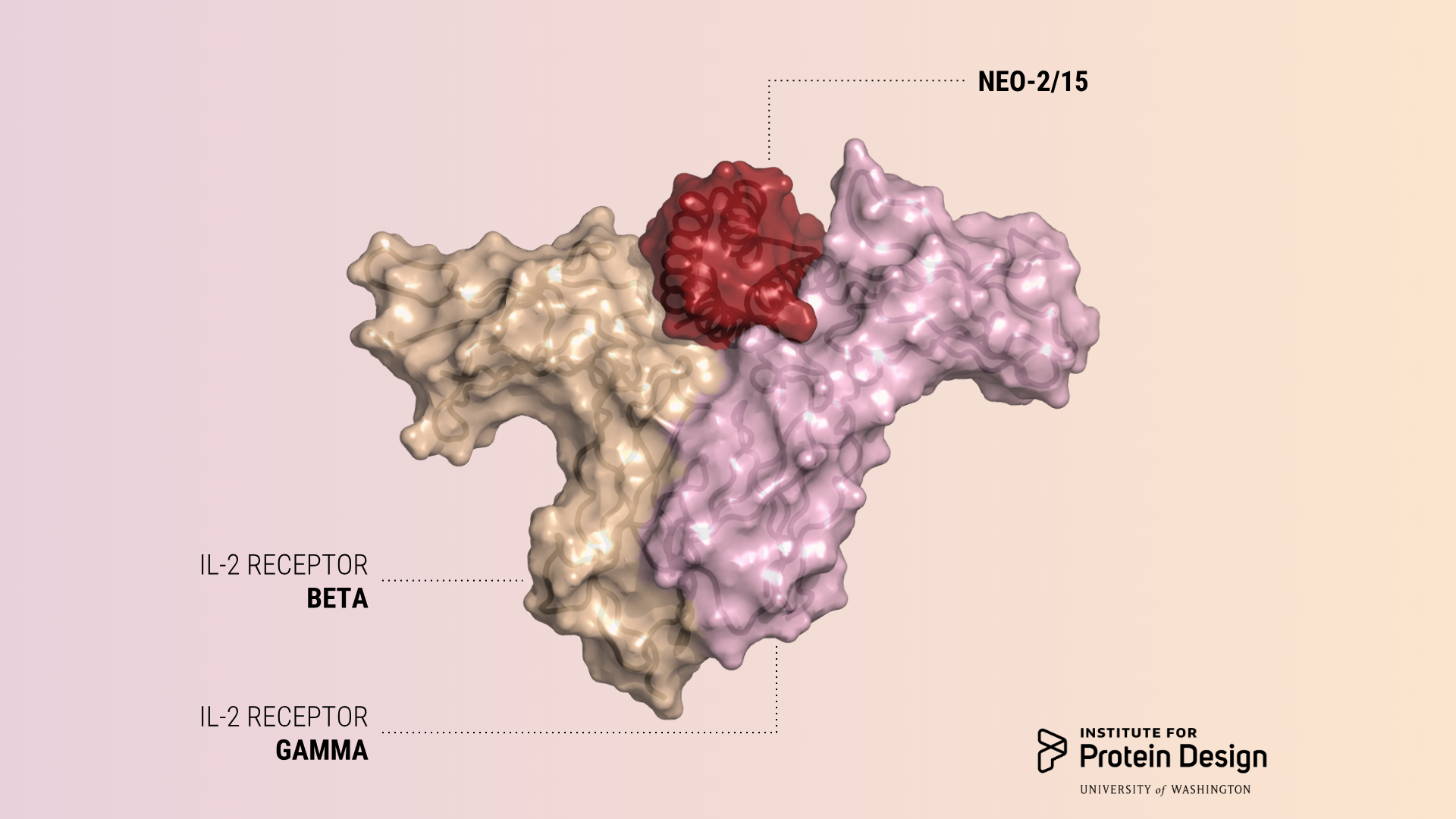
Potent anti-cancer proteins with fewer side effects
Today we report in Nature the first de novo designed proteins with anti-cancer activity. These compact molecules were designed to stimulate the same receptors as IL-2, a powerful immunotherapeutic drug, while avoiding unwanted off-target receptor interactions. We believe this is just the first of many computer-generated cancer drugs with enhanced specificity and potency. “People have tried for 30…
-
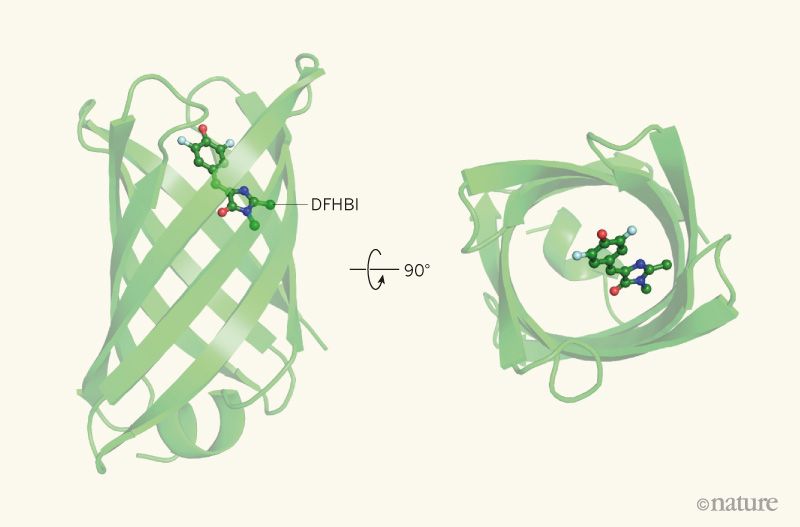
Our publication was voted ‘2018 Reader’s Choice’ by Nature News & Views
Readers of Nature News & Views selected an article about our work as their 2018 Reader’s Choice. The article, written by Roberto Chica of the University of Ottawa, details our recent publication on de novo fluorescence-activating proteins and explores the challenges of de novo protein design more generally. From the article: “The development and application…
-
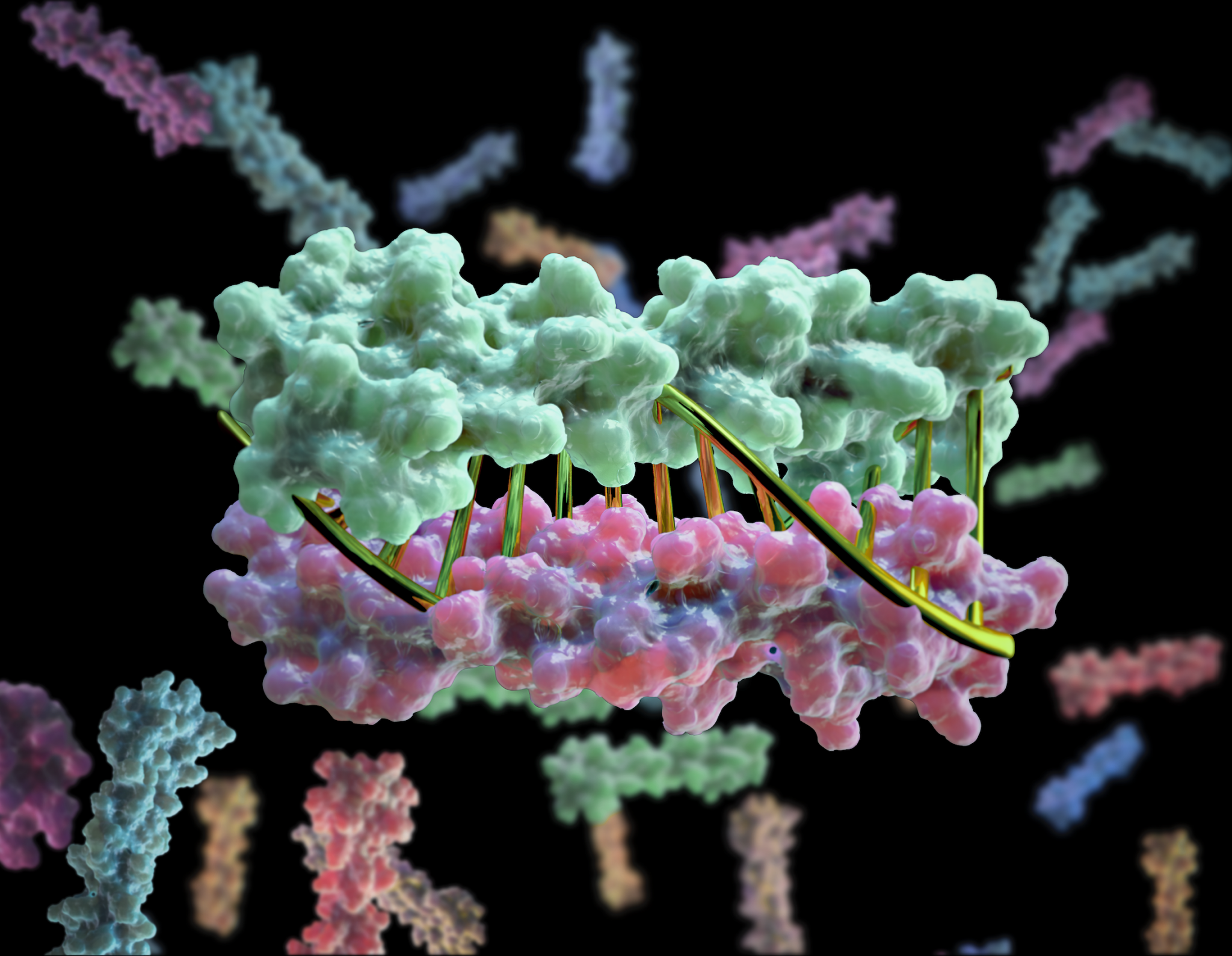
New designer proteins mimic DNA
To close out the year, Baker Lab scientists published a new report describing the creation of proteins that mimic DNA. We believe this breakthrough will aid the creation of bioactive nanomachines. DNA is a widely used building material at the nanoscale because it is simple and predictable: A pairs with T and C pairs with…
-
How synthetic biology could treat celiac disease
Dr. Ingrid Pultz, an IPD Translational Investigator and Chief Scientific Officer at PvP Biologics, has written a special report for the American Council on Science and Health about how protein design is being used to help fight celiac disease. Pultz describes how an international competition, a video game, and venture capital all aligned to help…
-
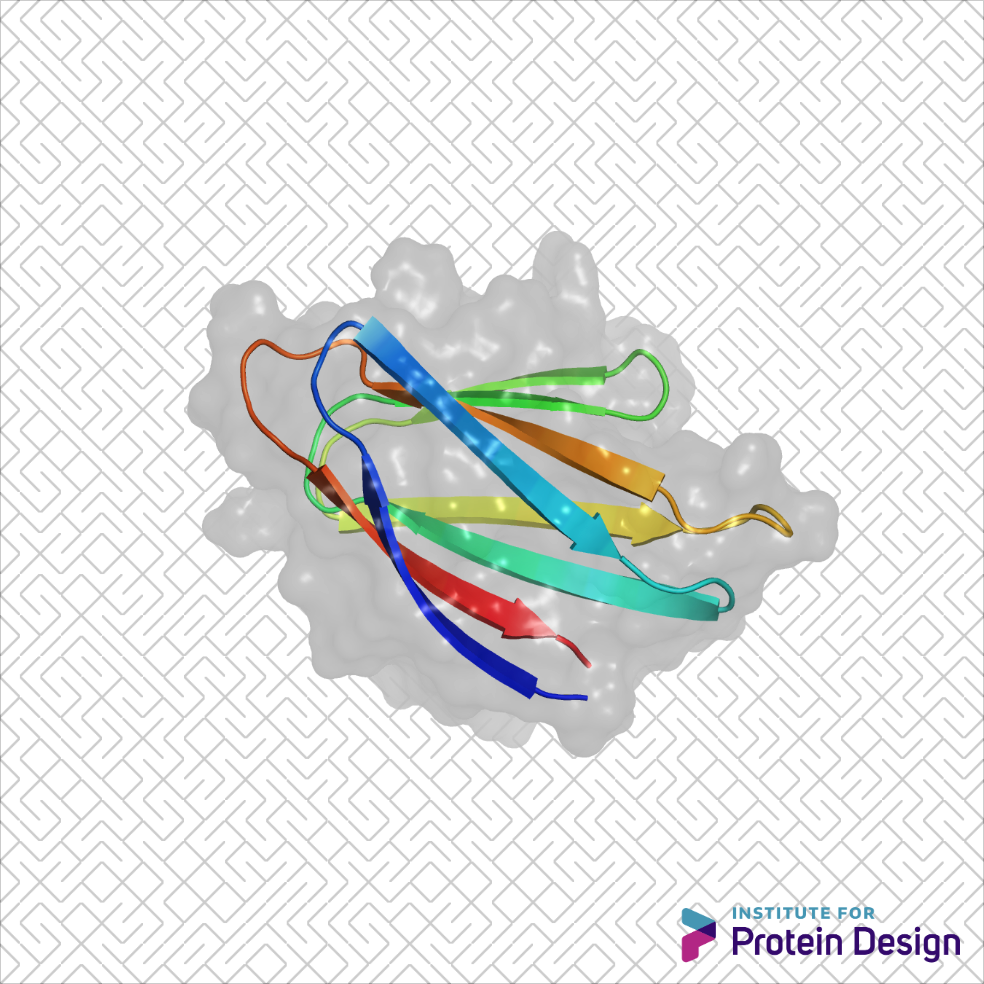
Rolling out new jellies
The basic parts of proteins — helices, strands and loops — can be combined in countless ways. But certain combinations are trickier than others. This week scientists from the IPD, along with collaborators in Brno and Santa Cruz, published the first-ever example of designed non-local beta-strand interactions. Beta-sheet proteins carry out critical functions in biology,…
-

Fluorescent proteins designed from scratch
In the summer of 1961, Osamu Shimomura drove across the country in a cramped station wagon to scoop jellyfish from the docks of Friday Harbor. He wanted to discover what made them glow. It took Shimomura and other biochemists more 30 years to find a full answer. By then, recombinant DNA technology allowed researchers to…
-

Open Philanthropy awards $11.3 million to the Institute for Protein Design
The funds will support our technological revolution in protein design and enable the development of a universal flu vaccine. The $11.3 gift is one of the largest made to date by the San Francisco-based philanthropy in support of science. It is also the first to go to UW Medicine. The gift comes in two parts:…
-

SCI-STEM Symposium 2018
Update 2018-07-26: The 2018 SCI-STEM Symposium was recently featured in an eLife article. The Institute for Protein Design at the University of Washington held its first ever symposium aimed at providing strategies to address diversity challenges in science, technology, engineering, and math (STEM). The Strategies for Cultivating Inclusion in STEM (SCI-STEM) symposium featured leading keynote…
-
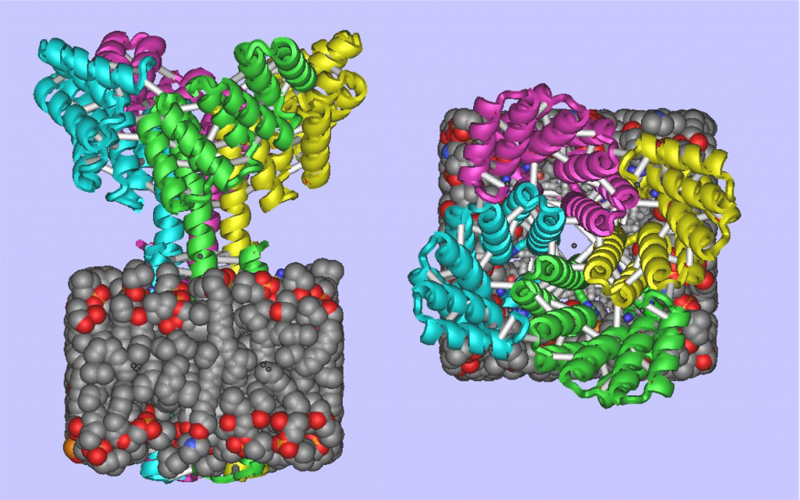
De Novo Design of Membrane Proteins
It is now possible to create complex, custom-designed transmembrane proteins from scratch ! Today Baker lab members published in Science “Accurate computational design of multipass transmembrane proteins” The Abstract reads as follows: The computational design of transmembrane proteins with more than one membrane-spanning region remains a major challenge. We report the design of transmembrane monomers,…
-
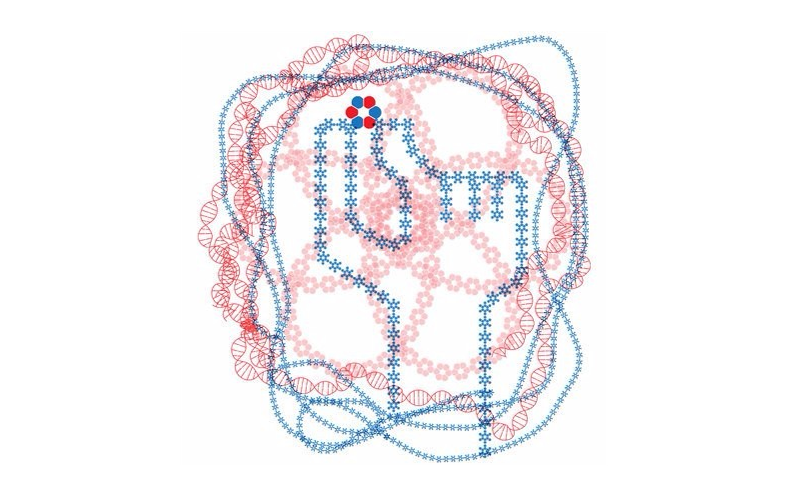
David Baker profiled in The New York Times
At the end of a historic year for protein design, the Baker Lab was honored to be profiled in the New York Times by science writer Carl Zimmer. He writes about the technology, progress, and promise in the field, including the contributions from our wonderful crowdsource participants. Graphic: John Hersey / New York Times On the…
-
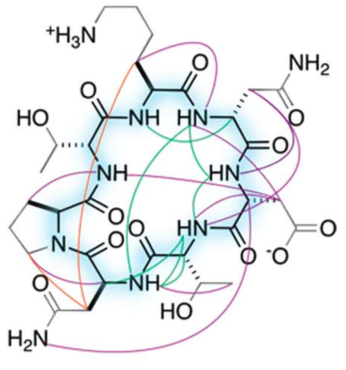
A New World of Designed Macrocycles
Today marks another major step forward for peptide based drug discovery. IPD researchers report in Science the computational design of a new world of small cyclic peptides, “Macrocycles”, increasing the number of the known kinds of these molecules by multiple fold. The conceptual art image below “Illuminating the energy landscape” shows the power of computational design…
-
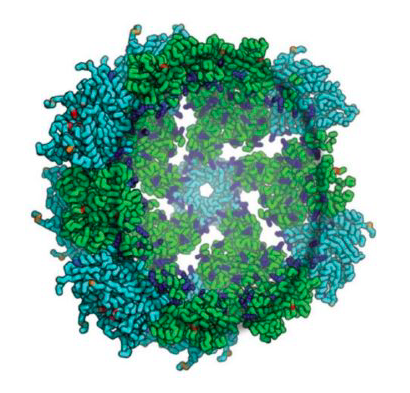
Synthetic Nucleocapsids Have Arrived
Published today in Nature, IPD researchers describe the first synthetic protein assemblies — dubbed synthetic nucleocapsids — that encapsulate their own genome and evolve in complex environments. Synthetic nucleocapsids are built to resemble viral capsids and could be used in future to deliver therapeutics to specific cells and tissues. These icosahedral protein assemblies are based off of…
-

Foldit to Disarm a Fungal Toxin
Foldit alfatoxin project update 7/16/2018 Today, scientists from of the Institute for Protein Design will join Foldit gamers from around the world to help design an enzyme that can neutralize aflatoxin — a cancer-causing toxin produced by certain fungi that are found on agricultural crops such as corn, peanuts, cottonseed, and tree nuts. Foldit is a citizen…
-

Designs on New World of Mini-Protein Therapeutics
Mark your calendars! September 27, 2017 is the day the doors opened to whole new world of targeted therapeutics. The Baker lab and numerous talented collaborators published in Nature that it is now possible to conduct “Massively parallel de novo protein design for targeted therapeutics”. Three factors make this possible: Rosetta molecular modeling algorithms for computational protein…
-
Cyrus Raises $8M to Advance Cloud-Based Protein Modeling and Design
Today, the first IPD spin out company Cyrus Biotechnology announced the closing of an $8M total Series A financing. The investment was led by Trinity Ventures, with participation from OrbiMed Advisors, SpringRock Ventures, W Fund, WRF Capital (a major supporter of the IPD), and individual investors. Congratulations Cyrus team! Cyrus is commercializing Cyrus Bench® an innovative user friendly software as…
-
Arzeda Raises $12M for Computational Protein Design
Today, Baker lab spin out company Arzeda announced that it had raised $12 million in a Series A round of funding led by OS Fund and including Bioeconomy Capital and Sustainable Conversion Ventures, as well as a follow-on investment from Arzeda’s seed investor, WRF Capital (a major supporter of the IPD). The new funding will enable “Technology…
-
The Matrix of Protein Design
The Matrix movie (1999) depicts a future in which the reality perceived by most humans is actually a computer simulated reality called “the Matrix”. Published today in Science, the Baker lab and collaborators report on a new kind of Matrix – a new reality for large scale computational protein design which can achieve massive data driven improvements in our ability to design highly stable,…
-
Stopping Influenza with Flu-Glue
Today, a multidisciplinary team of researchers at the University of Washington, Fred Hutch, and The Scripps Research Institute published in Nature Biotechnology the computational design of a trimeric influenza-neutralizing protein that binds extremely tightly to the H3 hemagglutinin of 1968 Hong Kong pandemic influenza virus (A/Hong Kong/X31/1968). It also cross-reacts with human relevant H1, H2 and…
-
Thank you, Bruce and Jeannie Nordstrom!
A few months ago, it was announced that the Institute for Protein Design is one of UW Medicine’s Priorities in their ACCELERATE campaign. We are grateful to have this support not only from UW Medicine, but also from donors who are contributing funds so that we may continue our work. One such gift came from…
-
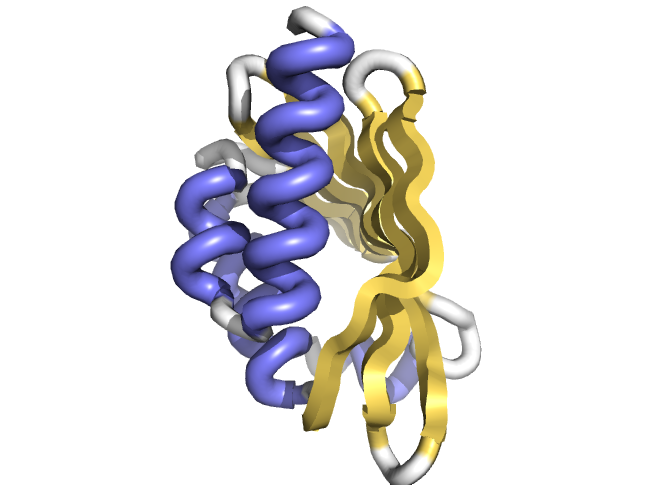
Design of novel cavity containing proteins
The latest paper coming out from the IPD was published today on the Science website. It’s titled “Principles for designing proteins with cavities formed by curved β sheets” with first co-authors Enrique Marcos and Benjamin Basanta, a former and current IPD member, respectively. Other IPD members on the paper include Tamuka Chidyausiku, Gustav Oberdorfer, Daniel-Adriano…
-
PvP Bio Announces $35M agreement with Takeda
On January 5th, recent IPD spin-out PvP Biologics announced their agreement with Takeda Pharmaceutical Company Limited. The $35 million deal includes an option to acquire PvP at a later point. PvP has released a statement on their website, and the agreement was highly covered by other news sources, which can be found at the…
-
New spinout: PvP Biologics!
We are happy to congratulate Ingrid Swanson Pultz, an IPD Translational Investigator, and Clancey Wolf, a Research Scientist, on PvP Biologics‘ spinout! The news was announced this morning and has been circulated through various outlets. The company, created in 2015, is focusing on advancing KumaMax, a gluten-fighting enzyme that could potentially be taken orally to…
-
Arzeda scales its automated molecule development pipeline
Seattle-based Arzeda, a computational and synthetic biology company that was spun out from the University of Washington labs of Prof. David Baker, recently announced that its high-throughput, automated pipeline for protein engineering and pathway discovery had been validated by the production of two keystone molecules. The announcement is a major technical milestone for Arzeda’s approach…
-

Unleashing the Power of Synthetic Proteins
Published today in Science Philanthropy Alliance, David Baker, Director of the Institute for Protein Design describes how the opportunities for computational protein design are endless — with new research frontiers and a huge variety of practical applications to be explored, from medicine to energy to technology. This is an exciting time as we are undergoing a technological revolution…
-
Limb Girdle Muscular Dystrophy Day and New Foldit puzzle
Today is Limb Girdle Muscular Dystrophy Day, and the Institute for Protein Design is collaborating with the Jain Foundation and Foldit community to to model the structure of human dysferlin protein (DYSF), an important protein for normal muscle function. Numerous mutations in the gene that encodes DYSF protein are known to be the cause of…
-

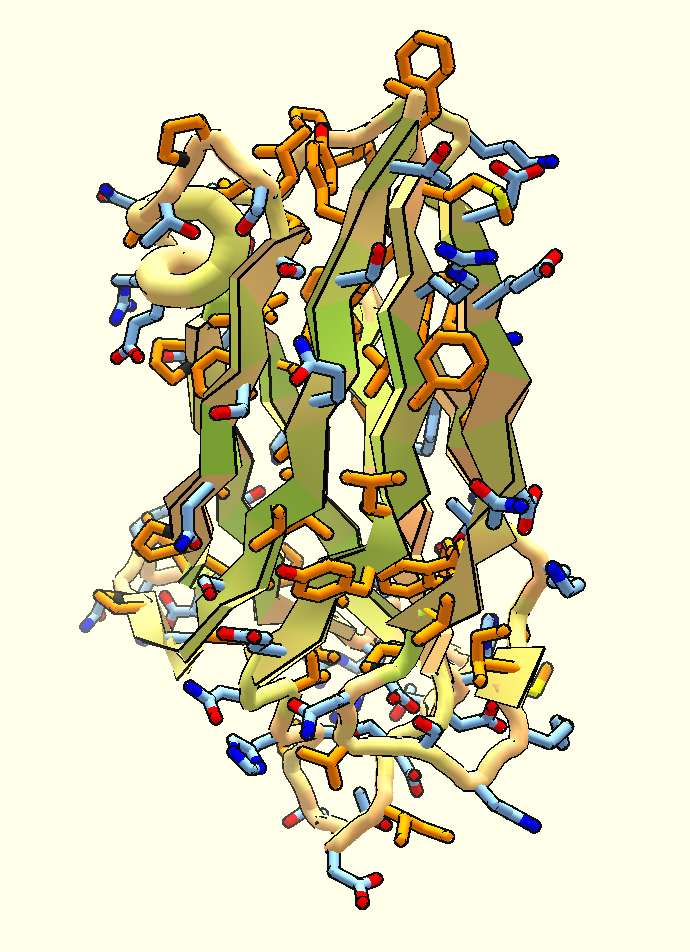
Hyper-stable Designed Peptides and the Coming of Age for De Novo Protein Design
Small constrained peptides combine the stability of small molecule drugs with the selectivity and potency of antibody-based therapeutics. However, peptide-based therapeutics have largely remained underexplored due to the limited diversity of naturally occurring peptide scaffolds, and a lack of methods to design them rationally. New computational design and wet lab methods developed at the Institute for…
-
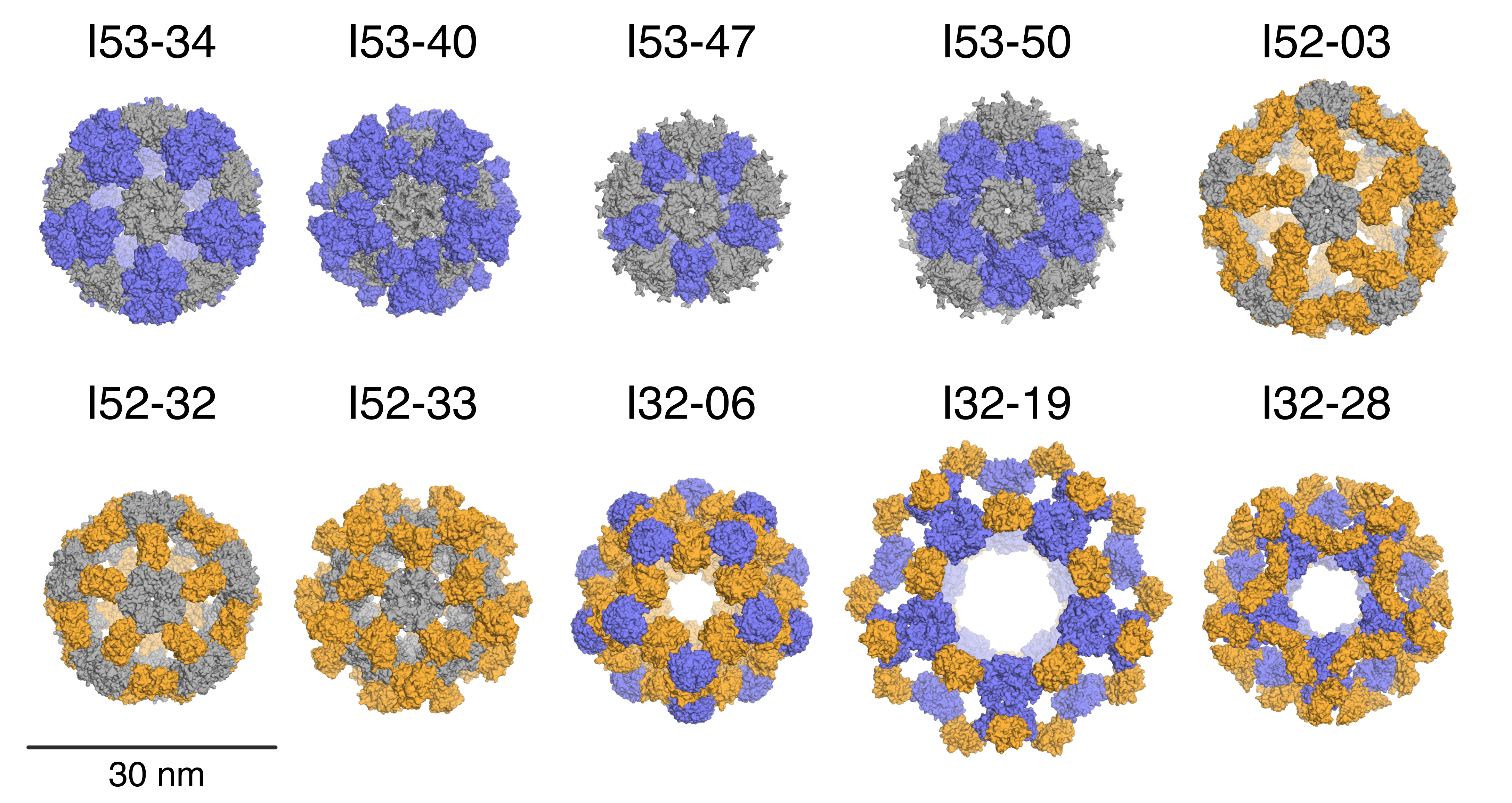
Designed Protein Containers Push Bioengineering Boundaries
Earlier this month, Baker lab researchers reported the computational design of a hyperstable 60-subunit protein icosahedron in Nature (Hsia et al); icosahedral protein structures are commonly observed in natural biological systems for packaging and transport (e.g. viral capsids). The described design was composed of 60 trimeric protein building blocks that self-assembled in a nanocage. In…
-
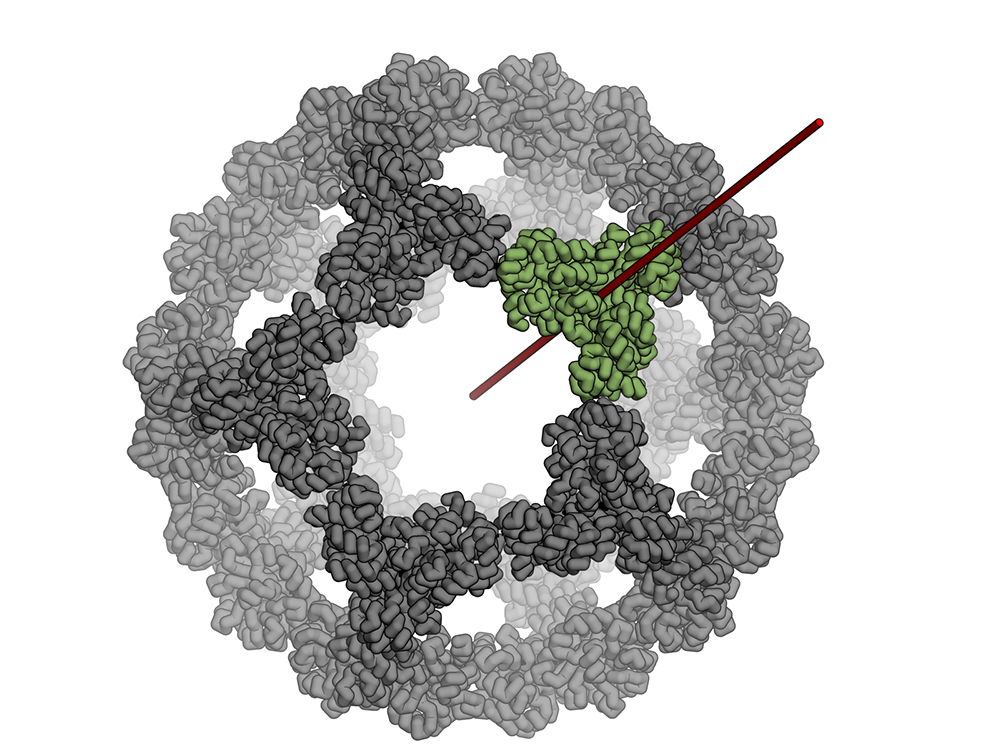
Icosahedral protein nanocage – new paper and podcast
The Baker lab, in collaboration with Neil King, Trisha Davis and Tamir Gonen’s labs, recently had a paper published in Nature about a stable icosahedral nanocage whose applications could span anywhere from drug delivery to vaccine design! The title is “Design of a hyperstable 60-subunit protein icosahedron” and it was published online June 15, 2016.…
-
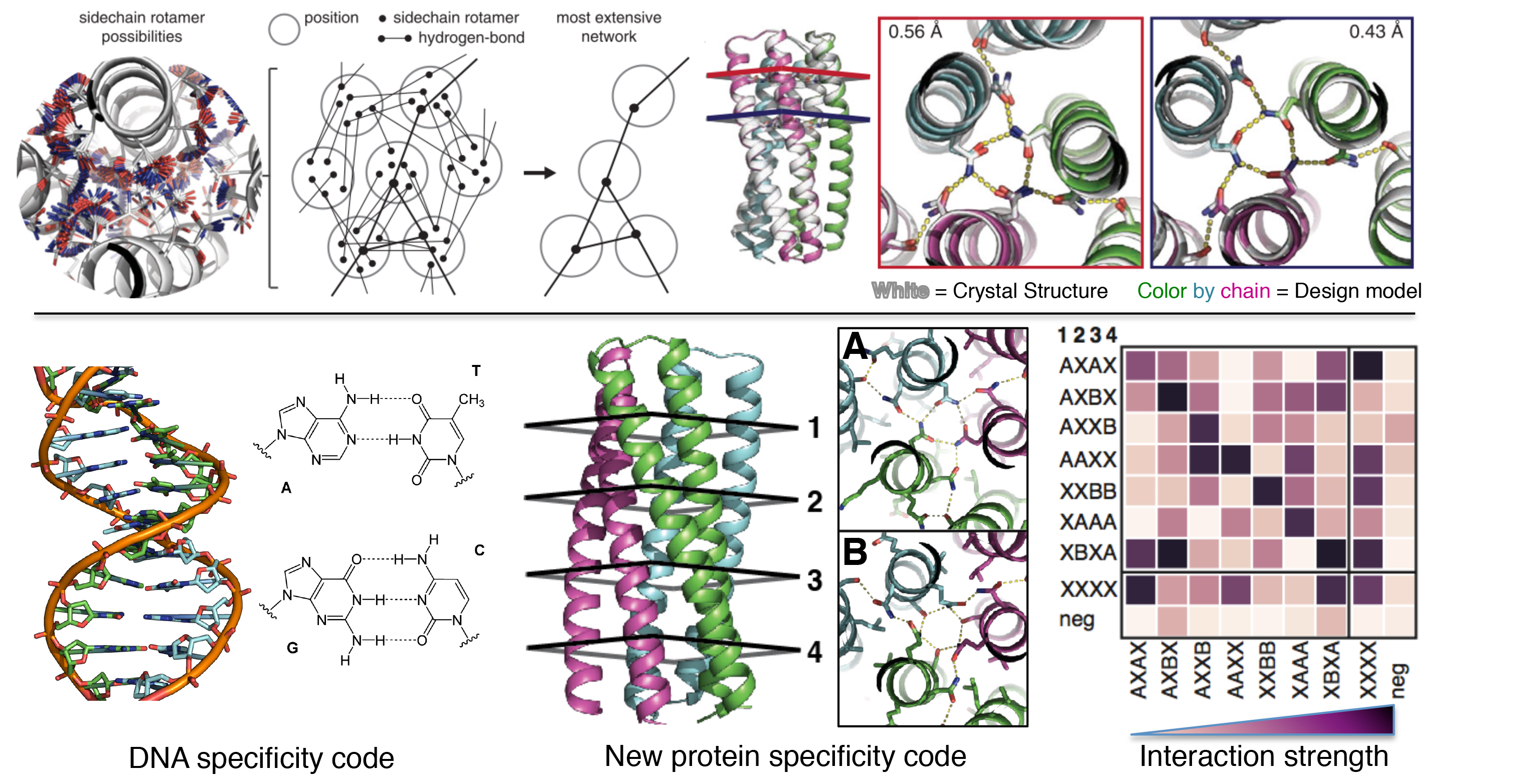
De novo design makes a splash
A paper recently published in Science by several members of the IPD, in collaboration with others, entitled “De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity,” discusses designing proteins in a similar way to DNA so that they may be used to engineer structures. Geekwire has written up a great article about the paper;…
-
Foldit Turns 8!
Over the weekend, Foldit had its 8th birthday! In celebration, they will be tweeting (@Foldit) fun facts and infographics on their feed. Haven’t heard of the game or tried playing it yet? What better time than now! Click here to learn more and to join an ever-growing community that spans the world. Who knows, maybe…
-

IPD at Xconomy’s EXOME’s Seattle’s Life Science Disruptors 2016
On Monday, representatives from the IPD spoke on a panel at Xconomy’s EXOME’s Seattle’s Life Science Disruptors 2016 event. It was titled “Proteins Like You’ve Never Seen” and included Lucas Nivon (Cyrus), Ingrid Swanson Putlz (PvP Biologics), Aaron Chevalier (Virvio), and David Baker. The event’s description was as follows: Seattle is one of the few…
-

David Baker wins a Leaders in Health Care Award, listed among ‘world’s most influential scientific minds’
We are proud to announce that last week, David was named winner of Seattle Business‘ 2016 Leaders in Health Care Awards for “Outstanding Achievement in Delivery of Digital Health.” The award was one of several given out, with categories ranging from “Lifetime Achievement” to “Outstanding Achievement: Health Care Delivery.” Although David was ill and unable…
-

Franziska Seeger @ Ignite Seattle
On February 18th, WRF Fellow Franziska Seeger gave a talk about proteins at Ignite Seattle. Presenters have 5 minutes to speak to the audience and deliver whatever point they are trying to get across. They also cannot control their slides, so they have to have their timing perfect. Talk about pressure! However, we believe that…
-

Arzeda and Mitsubishi Rayon Ltd announce new industrial collaboration
On February 9th, Arzeda, and Mitsubishi Rayon Ltd announced that they would “collaborate on development of new processes for industrial chemicals production.” Read more about it on Mitsubishi Rayon’s website, here, or on Arzeda’s site, here. Arzeda is an IPD spin-out, of which David Baker is a co-founder and Scientific Advisory Board Member.
-
Flu Binder Paper Debuts in PLOS Pathogens
Today at 11am, the paper titled “A Computationally Designed Hemagglutinin Stem-Binding Protein Provides In Vivo Protection from Influenza Independent of a Host Immune Response” was published to the PLOS Pathogen website. This paper was contributed to by several IPD members, including Aaron Chevalier, Jorgen Nelson, Lance Stewart, Lauren Carter, and David Baker. The research was performed…
-

Dr. Ingrid Swanson Pultz on UW Medicine Pulse
Recently, UW Medicine Pulse released a podcast featuring none other than our very own Ingrid Swanson Pultz! They talked to her about KumaMax and how it would help those with Celiac’s disease. Go here to see their post on it and listen to the podcast! Would you like to contribute to her research? Please go…
-
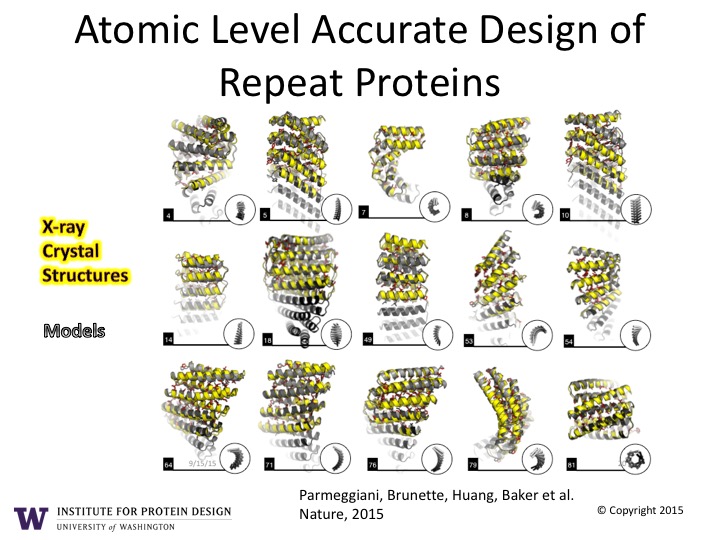
Big moves in protein structure prediction and design
[envira-gallery id=”3246″] Custom design with atomic level accuracy enables researchers to craft a whole new world of proteins Naturally occurring proteins are the nanoscale machines that carry out essentially all of the critical functions in living things. While it has been known for over 40 years that the sequence of amino acids completely determines the…
-
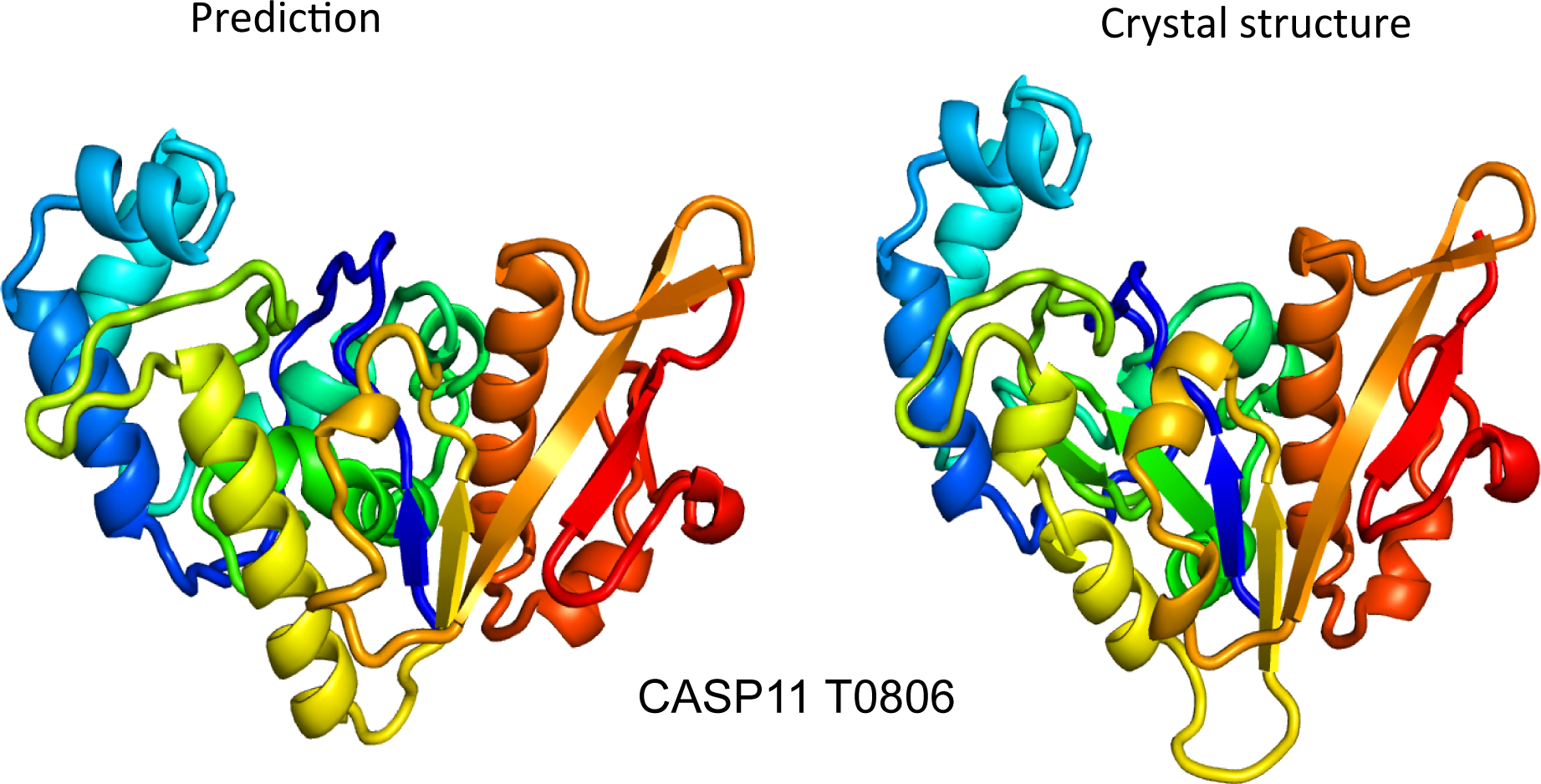
CASP3-11 Results Published in E-Life
In the early 1990s, researchers in the field of protein structure prediction were challenged by the problem of how to impartially judge the accuracy of prediction algorithms. This realization led the protein structure prediction the community to start the Critical Assessment of protein Structure Prediction (CASP), a community-wide, worldwide experiment for protein structure prediction taking…
-
September IPD News Roundup
RESEARCH IPD Translational Investigator, Dr. Ingrid Pultz, published a paper in JACS this month titled ‘Engineering of Kuma030: A Gliadin Peptidase That Rapidly Degrades Immunogenic Gliadin Peptides in Gastric Conditions‘. Using Rosetta to redesign the active site of the gliadin protease KumaMax – an enzyme computationally designed to break down gluten in the stomach – Dr. Pultz and…
-
Cyrus hires new EVP of Sales
Cyrus Biotechnology, the first company spinout from the IPD’s Translational Investigator Program, welcomes their new EVP of Sales, Rosario Caltabiano. Follow the link to the Cyrus website to learn more: http://cyrusbio.com/2015/10/01/cyrus-welcomes-evp-rosario-caltabiano/
-
August IPD News Roundup
RESEARCH Following the groundbreaking 2014 Nature paper describing the development of a computational method to design multi-component coassembling protein nanoparticles, comes a publication in Protein Science from Baker lab graduate student Jacob Bale and collaborators. Titled “Structure of a designed tetrahedral protein assembly variant engineered to have improved soluble expression“, the paper reports a variant of a previously low…
-
July IPD News Roundup
RESEARCH In an article out in Structure, Baker lab postdoc Dr. Hahnbeom Park in collaboration with IPD Assistant Professor Frank DiMaio investigate the origin of protein structure refinement from structural averaging at the residue level. Structure refinement has long been a challenge in the field of protein structure prediction and it aims to improve homology…
-
June IPD News Roundup
RESEARCH A new Science paper is out from Dr. David Baker and IPD collaborator Dr. Tamir Gonen (HHMI Janelia Campus) titled Design of ordered two-dimensional arrays mediated by noncovalent protein-protein interfaces. Graduate student Shane Gonen and Dr. Frank DiMaio describe a new computational approach to design two-dimensional protein arrays with a number of exciting potential applications…
-
New Science paper: Designed 2-D protein arrays
A new Science paper is out from IPD faculty Dr. David Baker titled Design of ordered two-dimensional arrays mediated by noncovalent protein-protein interfaces. Read the abstract below and the article at the link: http://www.sciencemag.org/content/348/6241/1365 We describe a general approach to designing two-dimensional (2D) protein arrays mediated by noncovalent protein-protein interfaces. Protein homo-oligomers are placed into one of the…
-
IPD Launches First Company Spinout
Seattle, WA Today, we are announcing that Cyrus Biotechnology, has been successfully launched from the UW Institute for Protein Design (IPD) to pursue commercialization of an innovative user friendly software as a service (SaaS) cloud computing solution for distribution of the powerful “Rosetta” protein structure prediction and design algorithms. “Cyrus is the first new IPD…
-
May IPD News Roundup
The May IPD News Roundup covers a new Science paper from IPD Assistant Prof Frank DiMaio, a KOMO News interview with Translational Investigator Ingrid Swanson Pultz on celiac disease, and much more! At the link.
-
New structure solved for hyperthermophilic DNA virus
A new Science paper is out from IPD faculty Dr. Frank DiMaio titled A virus that infects a hyperthermophile encapsidates A-form DNA. Read the abstract below and the article at the link: http://www.sciencemag.org/content/348/6237/914.full.pdf Extremophiles, microorganisms thriving in extreme environmental conditions, must have proteins and nucleic acids that are stable at extremes of temperature and pH. The nonenveloped,…
-
One-carbon pathway PNAS paper featured in Nature Chem Bio
The exciting protein design work by IPD researchers and collaborators in PNAS titled Computational protein design enables a novel one-carbon assimilation pathway has been featured in a Nature Chemical Biology News and Views piece. Follow the link to check it out: nchembio.1819
-
April IPD News Roundup
April IPD News Roundup is live at this link!
-
March IPD News Roundup
The March 2015 IPD news roundup is here! A new publication in PNAS on a completely novel metabolic pathway made via protein design and more. Read about it at the link.
-
February IPD News Roundup
This month’s roundup features an interview and two papers from IPD Assistant Professor Frank DiMaio as well as a new Science paper from the Baker lab on trapping transition states using protein design. Read more at the link.
-
AAAS 2015 Plenary Lecture by David Baker
David Baker, IPD Director and Professor of Biochemistry at the UW was the Plenary Speaker at this year’s AAAS meeting in San Francisco. A video of his talk, titled ‘Post-Evolutionary Biology: Design of Novel Protein Structures, Functions, and Assemblies’ covers a breadth of information on ongoing IPD research and can be viewed at the following…
-
January IPD News Roundup
A little late this month but it’s here!!! The January news roundup talks about a new Nature journal paper from the Baker lab, the second Mini Symposium, and much more. Follow the link and read more!
-
Independent Study Opportunity for Foster MBA Students
The Institute for Protein Design is seeking an MBA student to work with Institute leadership to collect market data on small molecule therapeutic targets for protein design. More information about this exciting independent study opportunity can be found on our Employment page here.
-
Institute for Protein Design Establishes Advisory Board
The Institute for Protein Design is pleased to announce its new advisory board! Follow the link to our news release for more information on our board members.
-
IPD Mini Symposium – Jan 20 2015
Our next Mini Symposium will be hosted on Jan 20 2015 and will feature Drs. Bill DeGrado (UCSF) and Gevorg Grigoryan (Dartmouth) speaking on “New Approaches to Protein Design”. More information at this link.
-
December IPD News Roundup
We wrapped up 2014 with a bang – introducing a new logo, getting a major award for celiac disease research, and successfully hosting our first Mini Symposium! Follow the link to read more.
-
Translational Investigator awarded LSDF Matching Grant Award for Celiac Disease Therapy
Please make a tax deductible donation at this LINK. IPD Translational Investigator Dr. Ingrid Swanson Pultz was awarded a Matching Grant award of $250K from the Life Sciences Discovery Fund (LSDF) for her project ‘In vivo assessment of an oral therapeutic for celiac disease‘! We need to raise an additional $74K…
-
November IPD News Roundup
Staff and scientists of the IPD and Foldit volunteered at Pacific Science Center’s Life Sciences Research Weekend this month – teaching budding scientists about the awesomeness of protein folding! Pics from the event and more Institute news at the link.
-
IPD Mini Symposium 2014
The UW Institute of Protein Design (IPD) presents a Mini Symposium on “Sweet Spots for Designed Proteins as Therapeutics” on Weds Dec 10 at 9 AM in HSB D-209. Follow the link to get more details! We hope to see you there!
-
October IPD News Roundup
Happy Fall from the IPD! Read about our October happenings in this month’s news roundup. Click here to read more.
-
Custom design of novel alphahelical bundles
A new paper is out in this week’s issue of Science entitled High thermodynamic stability of parametrically designed helical bundles. Using novel computational design methods, extremely stable helical bundles can be custom designed with fine-tuned structural geometries for a number of applications. Read more about this exciting work at this link.
-
September IPD News Roundup
We awarded our first round of WRF Innovation Postdoctoral fellowships this month! Follow the link to learn about our new fellows and to catch up on the research ongoing at the IPD.
-
August IPD News Roundup
The Foldit community has been leading the charge in the computational design of proteins to bind Ebola. Learn more about this work and other ongoing Institute news at the link.
-
Engineering an Oral Therapeutic for Celiac Disease
Learn how the IPD and Translational Investigator Dr. Ingrid Swanson Pultz are developing Kumamax, an oral therapeutic candidate for celiac disease. A slide presentation and more information at the link.
-
Targeting Ebola
The Foldit community to targets Ebola. Click here to learn more. Watch grad student Brian Koepnick and IPD Director David Baker talk about this work on this KOMO TV news segment. See the Seattle Times article here. Make a donation to help us fund our anti-Ebola effort.
-
Hiking the South Peak of The Brothers
Watch a video documenting a recent hike at The Brothers in the Olympics. IPD scientists, mountain goats, and stellar views – oh my!
-
The Power of Charity for Protein Design
Read a post about how the power of volunteer computing drives advances in protein design! Learn more at the link.
-
Letter from the Director – IPD Update
Read a letter from Institute for Protein Design Director Dr. David Baker summarizing the exciting accomplishments and progress made at the IPD in the past year.
-
July IPD News Roundup
This July, IPD staff and scientists ventured outdoors for a conference and for some fun adventures. Learn more at the link.
-
June IPD News Roundup
June at the IPD welcomed some new team members and saw some exciting new publications! Learn more here.
-
Designer Proteins to Target Cancer Cells
What if scientists could design a completely new protein that is precision-tuned to bind and inhibit cancer-causing proteins in the body? Collaborating scientists at the UW Institute for Protein Design (IPD) and Molecular Engineering and Sciences Institute (MolES) have made this idea a reality with the designed protein BINDI. BINDI (BHRF1-INhibiting Design acting Intracellularly) is a completely novel protein, based…


