The WHO estimates that venomous snakes kill over 100,000 people each year and leave 300,000 others with severe injuries, including lost limbs. This harm is particularly severe in regions that lack strong healthcare systems. Current antivenom therapies, derived from animal plasma, are expensive, difficult to produce, and carry risks of severe side effects.
In a new study published Nature, we introduce a new approach: AI-generated proteins that counteract deadly snake toxins. These custom toxin-binding proteins could pave the way for safer, more cost-effective, and widely available next-generation snakebite treatments.
Led by recent Baker Lab researcher Susana Vázquez Torres, this project included key collaborators from the Digital Biotechnology Lab at the Technical University of Denmark.
“I believe protein design will help make snake bite treatments more accessible for people in developing countries. The antitoxins we’ve created are easy to discover using only computational methods. They’re also cheap to produce and robust in laboratory tests.”
Susana Vázquez Torres, PhD
Collaborators on this project also included researchers from the Core R&D Labs at the UW Institute for Protein Design, University of Northern Colorado, Liverpool School of Tropical Medicine, Lancaster University, Sophion Bioscience, University of Liverpool, and Massachusetts Institute of Technology.
Targeting toxins
The team focused on toxins produced by elapid snakes, a group that includes cobras and mambas. These snakes deliver venom containing three-finger toxins, which cause paralysis and cell death by disrupting communication between nerves and muscles. Current antivenoms are often ineffective against these specific toxins.
By applying our latest protein design methods, Susana’s team created small, stable proteins capable of sticking to key regions of the toxins. When tested in the lab, these designed proteins neutralized multiple subtypes of three-finger toxins.
In mouse studies, the proteins provided complete protection from lethal neurotoxin exposure, achieving survival rates of 80-100% depending on the specific toxin and antitoxin protein.
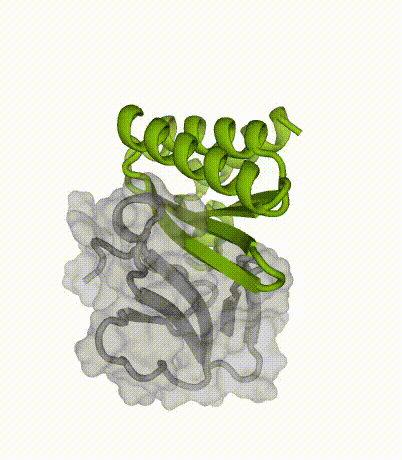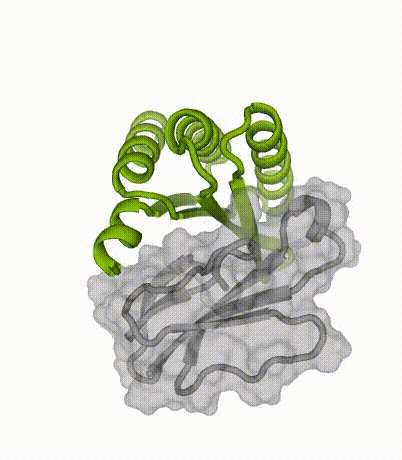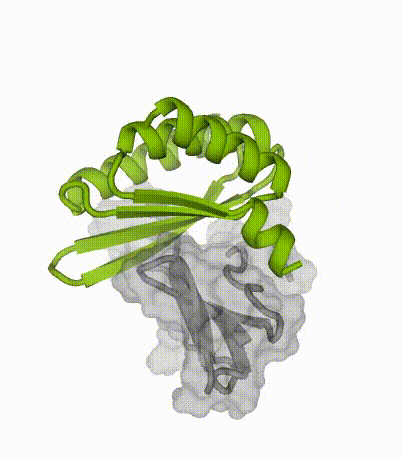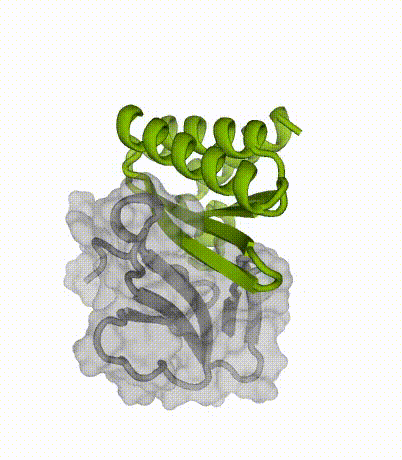
Advantages of miniproteins
“By designing binding proteins entirely on the computer using AI-powered software, we dramatically cut the time spent in the discovery phase,” explained co-senior author Tim Jenkins, an associate professor at DTU Bioengineering. “We didn’t need to perform several rounds of laboratory experiments to find proteins that performed well — the design software is so good now that we only needed to test a few molecules.”
Compact protein-binding proteins, also called miniproteins, may offer several advantages over traditional antivenoms:
- Recombinant Production: Miniproteins can be manufactured without the need for animal immunization, ensuring consistent quality.
- Smaller Size: Their compact structure may enable deeper tissue penetration for faster toxin neutralization.
- Thermal Stability: Like most computationally designed proteins, miniproteins tend to remain folded and active at high temperatures, potentially allowing for easier transport and longer storage.
The new proteins also showed no side effects in animal studies, a critical step toward safe deployment in human therapies.
Better medicines by design
Although traditional antivenoms will remain central to treating snakebite victims for the foreseeable future, these new computer-designed antitoxins could supplement existing treatments or enhance their effectiveness. Moreover, the same design approach can extend beyond snakebites to treat other diseases that lack effective therapies, including viral infection, autoimmune disease, and many other health disorders.
“Beyond treating snakebites, protein design will help simplify and democratize drug discovery, particularly in resource-limited settings,” said David Baker. “By lowering costs and resource requirements for protein-based medicines, we’re taking considerable steps toward a future where everyone can get the treatments they deserve.”
Coverage
AI designs ‘breakthrough’ snakebite treatment that could turbocharge antivenom development — The Telegraph
AI-designed proteins tackle century-old problem — making snake antivenoms — Nature
Funding
This work was funded by the Open Philanthropy Project, Audacious Project, Washington State General Operating Fund, Howard Hughes Medical Institute, Novo Nordisk Foundation, EuroTech Universities Alliance, European Union’s Horizon 2020 programme, Wellcome Trust, and the National Science Foundation. Support was provided by the Liverpool Shared Research Facilities, National Synchrotron Light Source II, and the Center for BioMolecular Structure. Crystallographic data collection was supported by NIH and Department of Energy grants. All funders are listed in the open-access manuscript.