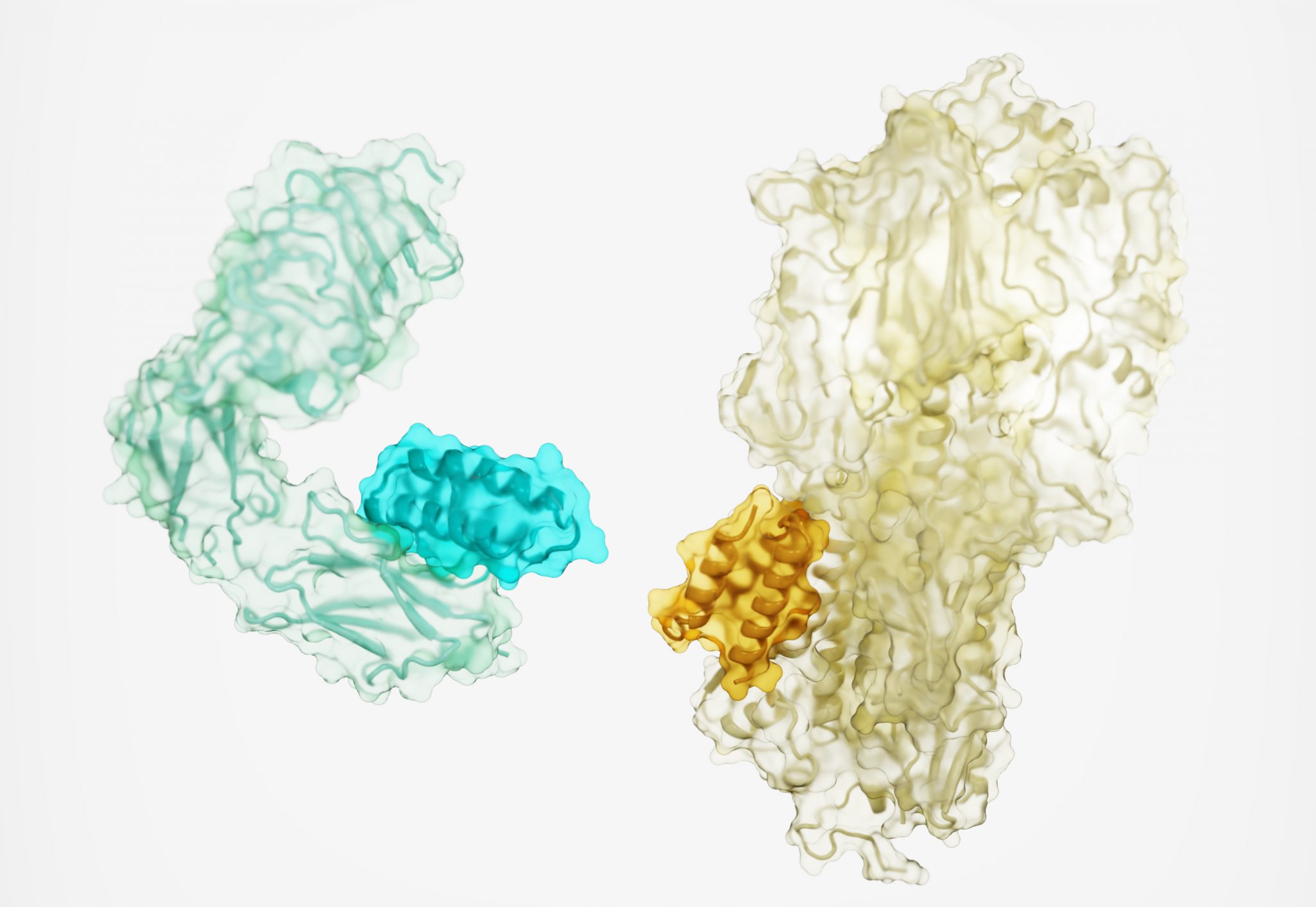
Today we report in Nature a new method for generating protein drugs. Using Rosetta-based design, an international team designed molecules that can target important proteins in the body, such as the insulin receptor, as well as proteins on the surface of viruses. This solves a long-standing challenge in drug development and may lead to new treatments for cancer, diabetes, infection, inflammation, and beyond.
This work was led by two Baker lab postdoctoral scholars – Longxing Cao, PhD, and Brian Coventry, PhD – who combined recent advances in computational protein design to arrive at a strategy for creating new proteins that bind molecular targets in a manner similar to antibodies. They developed software that can scan a target molecule, identify potential binding sites, generate proteins targeting those sites, and then screen from millions of candidate binding proteins to identify those most likely to function.
The team generated high-affinity binding proteins against 12 distinct molecular targets. These targets include important cellular receptors such as TrkA, EGFR, Tie2, and the insulin receptor, as well proteins on the surface of the influenza virus and SARS-CoV-2 (the virus that causes COVID-19).
“When it comes to creating new drugs, there are easy targets and there are hard targets,” said Cao, who is now an assistant professor at Westlake University. “In this paper, we show that even very hard targets are amenable to this approach. We were able to make binding proteins to some targets that had no known binding partners or antibodies.”
In total, the team produced over half a million candidate binding proteins for the 12 selected molecular targets. Data collected on this large pool of candidate binding proteins was used to improve the overall method.
“We look forward to seeing how these molecules might be used in a clinical context, and more importantly how this new method of designing protein drugs might lead to even more promising compounds in the future,” said Coventry.
The research team included scientists from the University of Washington School of Medicine, Yale University School of Medicine, Stanford University School of Medicine, Ghent University, The Scripps Research Institute, and the National Cancer Institute, among other institutions.
This work was supported in part by The Audacious Project at the Institute for Protein Design, Open Philanthropy Project, National Institutes of Health (HHSN272201700059C, R01AI140245, R01AI150855, R01AG063845), Defense Advanced Research Project Agency (HR0011835403 contract FA8750-17-C-0219), Defense Threat Reduction Agency (HDTRA1-16-C-0029), Schmidt Futures, Gates Ventures, Donald and Jo Anne Petersen Endowment, and an Azure computing gift for COVID-19 research provided by Microsoft.