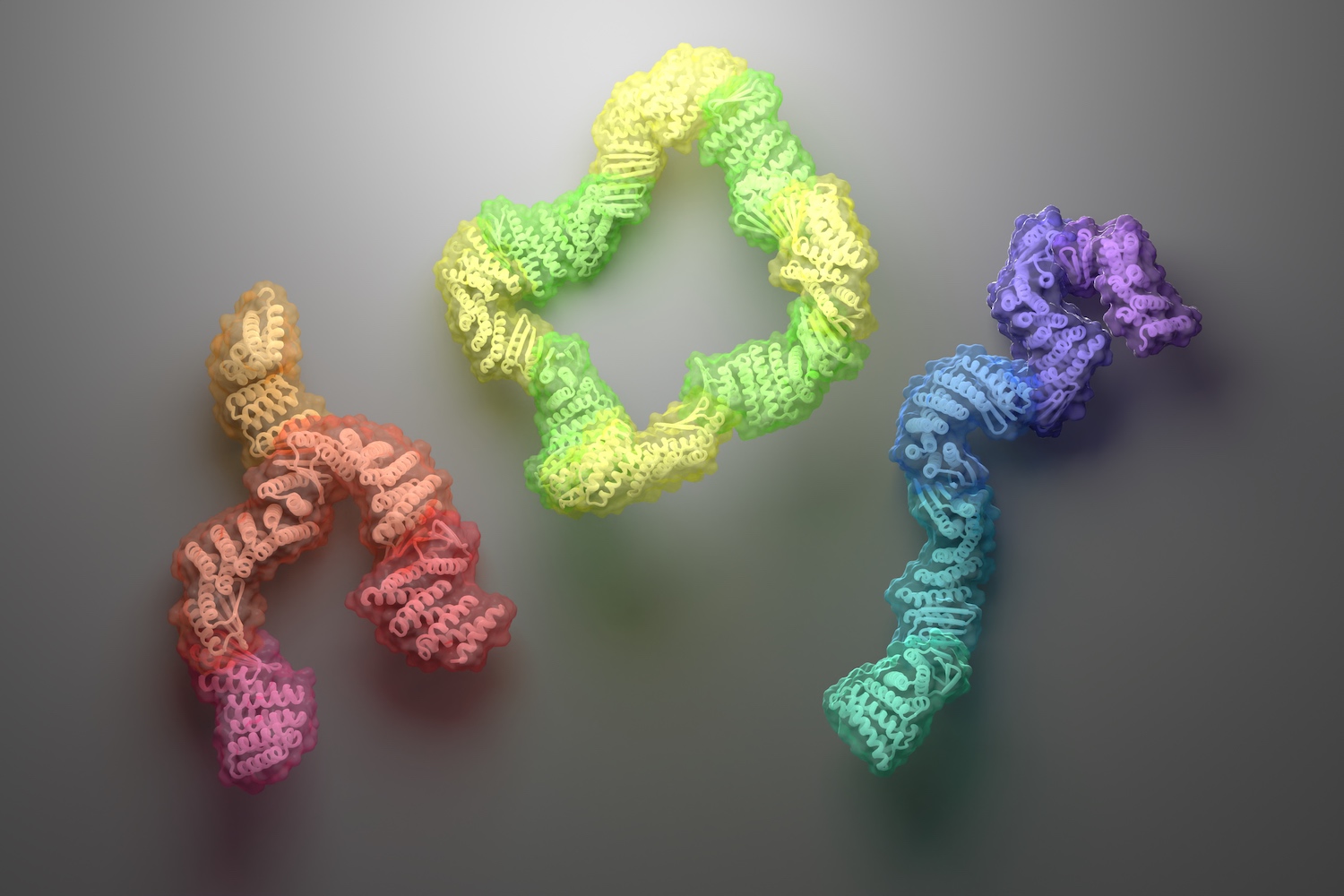A new approach for creating custom protein complexes yields asymmetric assemblies with interchangeable parts.
Today we report in Science the design of new protein assemblies made from modular parts. These complexes — which adopt linear, branching, or closed-loop architectures — contain up to six unique proteins, each of which remains folded and soluble in the absence of any binding partners. Baker lab postdoctoral scholars Danny Sahtoe and Florian Praetorius led this work.
Proteins in living cells often come together to form complexes that carry out vital functions. A key feature of these sophisticated assemblies is their ability to exchange parts as needed. To replicate DNA, for example, dozens of unique proteins in a cell spontaneously form into clamps, clamp-loading assemblies, and multi-chain enzyme complexes.
“We see many areas in synthetic biology that would benefit from the ability to pair up proteins in structurally defined ways, but there is currently no easy way to do this,” said Praetorius. “For this project, we wanted to create stable proteins that could spontaneously and rapidly assemble into predictable configurations upon mixing. These could then be fused to other molecules to drive them together.”

Implicit negative design
To begin, the team first sought to create new pairs of proteins that would bind reliably to their cognate partners but not themselves. Such stable heterodimers could then serve as junction points for larger multi-protein architectures.
“To create stable heterodimers, we turned to negative design,” explains Sahtoe. “We decided to introduce three properties into our proteins that make self-association unlikely. First, we made our proteins stable by including real hydrophobic cores. Second, we designed interfaces that require two beta-sheets to pair up, creating unique and continuous beta-sheets across the heterodimer interfaces. And finally, we used Rosetta to model the possible unwanted homooligomeric states and then used that information to prevent those states.”
Using these design principles, the team generated several new heterodimeric proteins that assemble rapidly, even in crowded cell lysates. High-resolution crystal structures of three such dimers confirmed the intended binding modes.
These well-behaved proteins were then recombined and fused to create new “connector proteins” that could bind partners at either end. The team went on to create branched, star-shaped, and angled connectors as well. These modular parts can in principle be mixed and matched to create roughly 500 unique multi-chain assemblies.
The new self-assembling protein parts function as designed in living cells, mediating the assembly of liquid-liquid condensates or more solid assemblies depending on the interaction affinities. The team also showed that assemblies that had already formed could be reconfigured by mixing in new components.
This work was supported by EMBO long-term fellowships, Human Frontiers Science Program long-term fellowships, a Washington Research Foundation Innovation fellowship, and by NIH, DARPA, HHMI, Open Philanthropy Project, The Audacious Project, and Eric and Wendy Schmidt by recommendation of the Schmidt Futures. All data are available in the main text or supplementary materials. Design scripts, protein sequences, design models, and models of assemblies are also available through Zenodo.