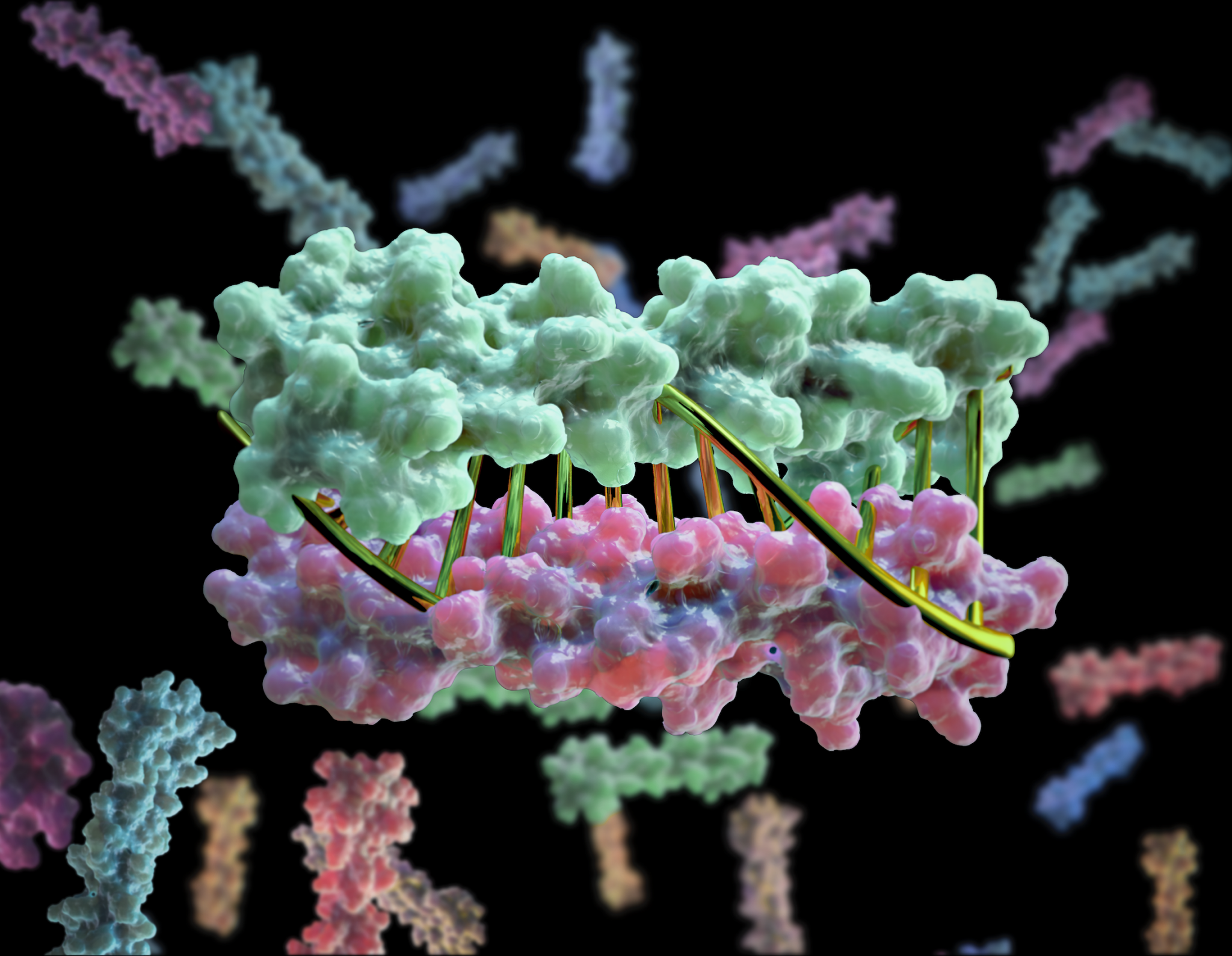
To close out the year, Baker Lab scientists published a new report describing the creation of proteins that mimic DNA. We believe this breakthrough will aid the creation of bioactive nanomachines.
DNA is a widely used building material at the nanoscale because it is simple and predictable: A pairs with T and C pairs with G. Because of this, DNA strands can be programmed to click together into precise and increasingly complex structures. But DNA has drawbacks. It is not as bioactive as RNA, and not nearly as active as proteins. Bioactive protein assemblies run cells (kinetochores, polymerases, proteasomes, etc). What if designing them was as easy as clicking together DNA?
Using computational design, we created heterodimeric proteins that form double helices with hydrogen-bond mediated specificity. When a pool of these new protein zippers gets melted and then allowed to refold, only the proper pairings form. They are all-against-all orthogonal. With these new tools in hand, we can now begin constructing large protein-based machines that self-assemble in predictable ways.
This project was led by graduate student Zibo Chen and was done in collaboration with the Wysocki Lab at Ohio State University and the Sgourakis Lab at the UC Santa Cruz. The work used support from the SIBYLS program with SAXS and the ALS resources at LBNL, as well as the Argonne Leadership Computing Facility at ANL.
Read the full here: https://www.nature.com/articles/s41586-018-0802-y (PDF)