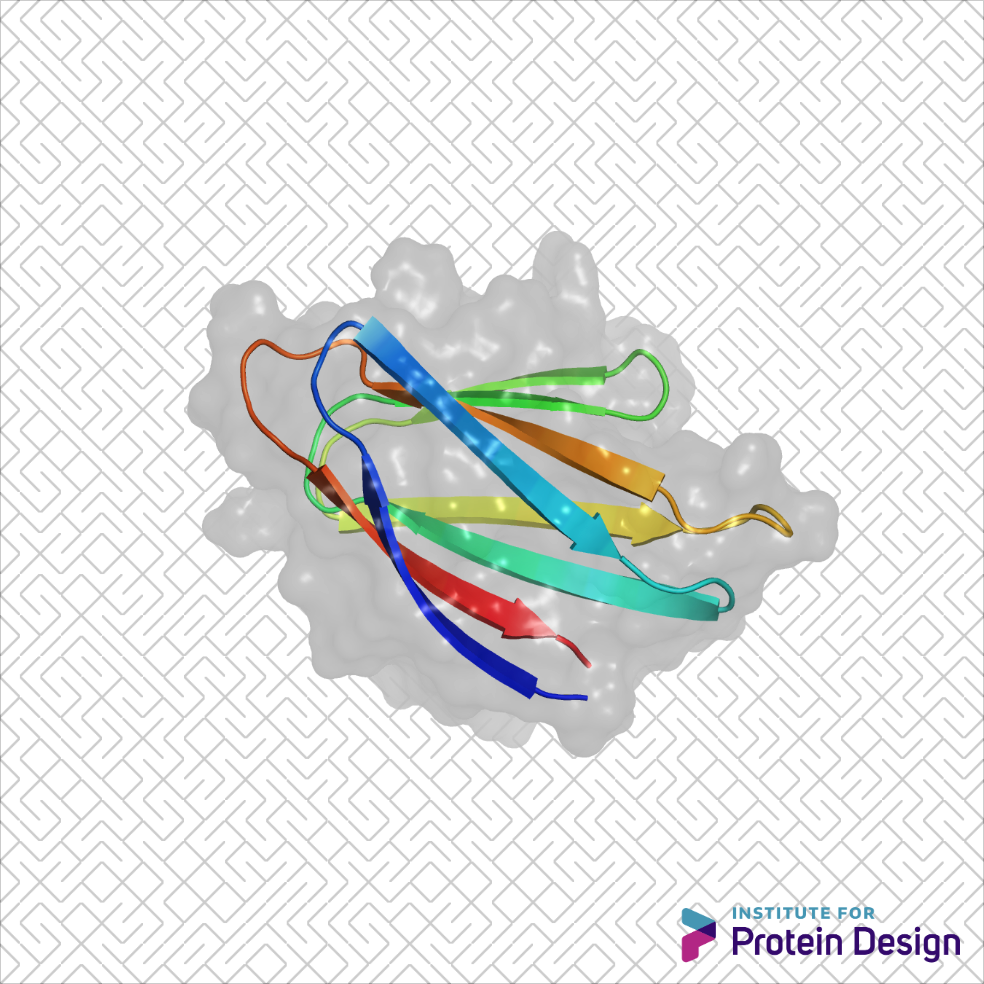
The basic parts of proteins — helices, strands and loops — can be combined in countless ways. But certain combinations are trickier than others. This week scientists from the IPD, along with collaborators in Brno and Santa Cruz, published the first-ever example of designed non-local beta-strand interactions.
Beta-sheet proteins carry out critical functions in biology, and hence are attractive scaffolds for computational protein design, but the de novo design of all-beta-sheet proteins from first principles has lagged far behind the design of all-alpha or mixed-alpha-beta domains.

Local beta-strand interactions occur when residues near one other hydrogen bond to form compact sheets. To get similar interactions from stretches of residues that are not close in primary sequence, a protein backbone must fold into a complex interwoven shape. The successful design of non-local beta-strand interactions demonstrates a significant advance in our ability to control both fine features (such as precise hydrogen bonding) and global features (such as complex topology) in proteins and opens the door to the design of a broad range of non-local beta-sheet structures.
By studying loops that connect unpaired beta-strands (beta-arches), the team identified a series of structural relationships between loop geometry, side chain directionality and beta-strand length that arise from hydrogen bonding and packing constraints on regular beta-sheet structures. They used these rules to de novo design jellyroll structures with double-stranded beta-helices formed by eight antiparallel β-strands. NMR of a hyperthermostable design closely matched the computational model, demonstrating accurate control over the beta-sheet structure and loop geometry.
Read the full report here: https://www.nature.com/articles/s41594-018-0141-6 (PDF)