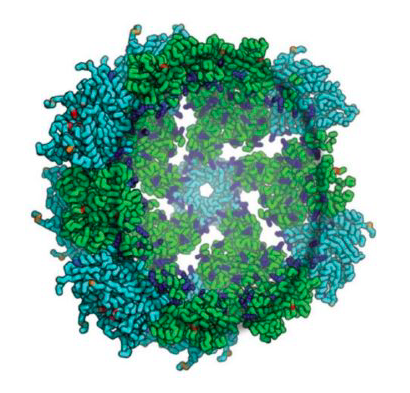
Published today in Nature, IPD researchers describe the first synthetic protein assemblies — dubbed synthetic nucleocapsids — that encapsulate their own genome and evolve in complex environments.

Synthetic nucleocapsids are built to resemble viral capsids and could be used in future to deliver therapeutics to specific cells and tissues. These icosahedral protein assemblies are based off of previously reported results from the Institute for Protein Design.
The image above visualizes the de novo creation of synthetic nucleocapsids from computationally designed proteins and their evolution to acquire properties that could be useful for drug delivery and other biomedical applications. The narrative was designed as a futuristic hologram projection realized through spiraling DNA composed of binary zeros and ones. The projection and computational imagery evoke futuristic technology and design, while calling out natural evolution through the DNA spiral “time-scale” motif. The heads-up display iconography showing a blood bag, mouse, and RNase A convey the challenges we used to evolve the synthetic nucleocapsids. The single net impression of this image is engaging + enlightening and shows that we are entering the next epoch of synthetic biology in which biological systems can be designed and created from scratch.
Abstract:
The challenges of evolution in a complex biochemical environment, coupling genotype to phenotype and protecting the genetic material, are solved elegantly in biological systems by the encapsulation of nucleic acids. In the simplest examples, viruses use capsids to surround their genomes. Although these naturally occurring systems have been modified to change their tropism and to display proteins or peptides, billions of years of evolution have favoured efficiency at the expense of modularity, making viral capsids difficult to engineer. Synthetic systems composed of non-viral proteins could provide a ‘blank slate’ to evolve desired properties for drug delivery and other biomedical applications, while avoiding the safety risks and engineering challenges associated with viruses. Here we create synthetic nucleocapsids, which are computationally designed icosahedral protein assemblies with positively charged inner surfaces that can package their own full-length mRNA genomes. We explore the ability of these nucleocapsids to evolve virus-like properties by generating diversified populations using Escherichia coli as an expression host. Several generations of evolution resulted in markedly improved genome packaging (more than 133-fold), stability in blood (from less than 3.7% to 71% of packaged RNA protected after 6 hours of treatment), and in vivo circulation time (from less than 5 minutes to approximately 4.5 hours). The resulting synthetic nucleocapsids package one full-length RNA genome for every 11 icosahedral assemblies, similar to the best recombinant adeno-associated virus vectors. Our results show that there are simple evolutionary paths through which protein assemblies can acquire virus-like genome packaging and protection. Considerable effort has been directed at ‘top-down’ modification of viruses to be safe and effective for drug delivery and vaccine applications; the ability to design synthetic nanomaterials computationally and to optimize them through evolution now enables a complementary ‘bottom-up’ approach with considerable advantages in programmability and control.
Read more and obtain a PDF of the paper from the Baker lab web site. Also read additional news items from Science Daily, GeekWire, UW Newsroom, CEN.