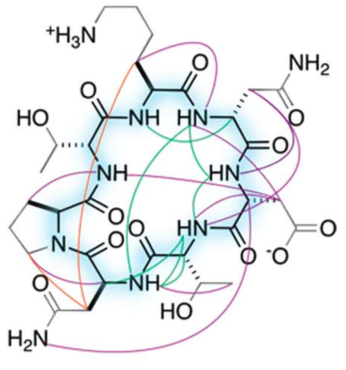
Today marks another major step forward for peptide based drug discovery. IPD researchers report in Science the computational design of a new world of small cyclic peptides, “Macrocycles”, increasing the number of the known kinds of these molecules by multiple fold. The conceptual art image below “Illuminating the energy landscape” shows the power of computational design to explore and illuminate structured peptides across the vast energy landscape.
Small peptides have the benefits of small molecule drugs, like aspirin, and large antibody therapies, like rituximab, with fewer drawbacks. They are stable like small molecules and potent and selective like antibodies.
Abstract reads as follows.
Mixed-chirality peptide macrocycles such as cyclosporine are among the most potent therapeutics identified to date, but there is currently no way to systematically search the structural space spanned by such compounds. Natural proteins do not provide a useful guide: Peptide macrocycles lack regular secondary structures and hydrophobic cores, and can contain local structures not accessible with L-amino acids. Here, we enumerate the stable structures that can be adopted by macrocyclic peptides composed of L- and D-amino acids by near-exhaustive backbone sampling followed by sequence design and energy landscape calculations. We identify more than 200 designs predicted to fold into single stable structures, many times more than the number of currently available unbound peptide macrocycle structures. Nuclear magnetic resonance structures of 9 of 12 designed 7- to 10-residue macrocycles, and three 11- to 14-residue bicyclic designs, are close to the computational models. Our results provide a nearly complete coverage of the rich space of structures possible for short peptide macrocycles and vastly increase the available starting scaffolds for both rational drug design and library selection methods.
Check out these additional news items from UW Medicine, and Science.
Read more and download the paper at the Baker lab web site.